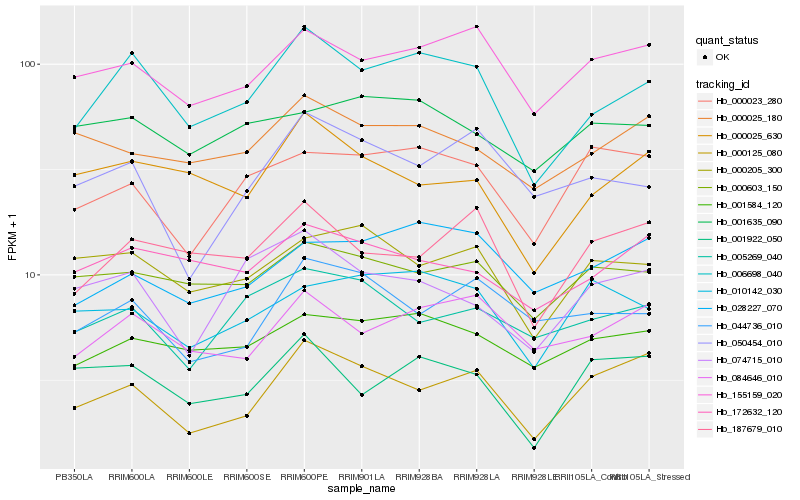
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006698_040 |
0.0 |
- |
- |
bHLH1 transcription factor [Hevea brasiliensis] |
| 2 |
Hb_084646_010 |
0.1076947844 |
- |
- |
hypothetical protein CICLE_v10031746mg [Citrus clementina] |
| 3 |
Hb_001584_120 |
0.1093589898 |
- |
- |
PREDICTED: probable Xaa-Pro aminopeptidase 3 [Jatropha curcas] |
| 4 |
Hb_155159_020 |
0.1097541718 |
- |
- |
PREDICTED: clathrin light chain 1-like [Jatropha curcas] |
| 5 |
Hb_000025_630 |
0.1147477538 |
- |
- |
PREDICTED: uncharacterized protein LOC105635952 [Jatropha curcas] |
| 6 |
Hb_187679_010 |
0.1168148496 |
- |
- |
- |
| 7 |
Hb_172632_120 |
0.1183667498 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000205_300 |
0.1215966232 |
- |
- |
PREDICTED: uncharacterized protein LOC105647767 [Jatropha curcas] |
| 9 |
Hb_000603_150 |
0.1220220268 |
- |
- |
PREDICTED: guanine nucleotide-binding protein alpha-1 subunit [Jatropha curcas] |
| 10 |
Hb_028227_070 |
0.1254020804 |
- |
- |
PREDICTED: uncharacterized protein LOC105644820 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000025_180 |
0.1258384252 |
- |
- |
hypothetical protein CISIN_1g024241mg [Citrus sinensis] |
| 12 |
Hb_074715_010 |
0.1286245754 |
- |
- |
PREDICTED: disease resistance protein RPS6-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_001922_050 |
0.1294506912 |
- |
- |
PREDICTED: F-box protein At5g52880 [Jatropha curcas] |
| 14 |
Hb_000125_080 |
0.1305189378 |
transcription factor |
TF Family: mTERF |
- |
| 15 |
Hb_005269_040 |
0.1306500447 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: pentatricopeptide repeat-containing protein At1g06270 [Jatropha curcas] |
| 16 |
Hb_010142_030 |
0.1318848391 |
- |
- |
PREDICTED: isoamylase 3, chloroplastic isoform X2 [Jatropha curcas] |
| 17 |
Hb_001635_090 |
0.1325270283 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 7 isoform X1 [Populus euphratica] |
| 18 |
Hb_050454_010 |
0.1325329813 |
- |
- |
PREDICTED: uncharacterized protein LOC105630497 [Jatropha curcas] |
| 19 |
Hb_044736_010 |
0.1333240468 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 20 |
Hb_000023_280 |
0.1337427825 |
- |
- |
hypothetical protein POPTR_0006s28210g [Populus trichocarpa] |