| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
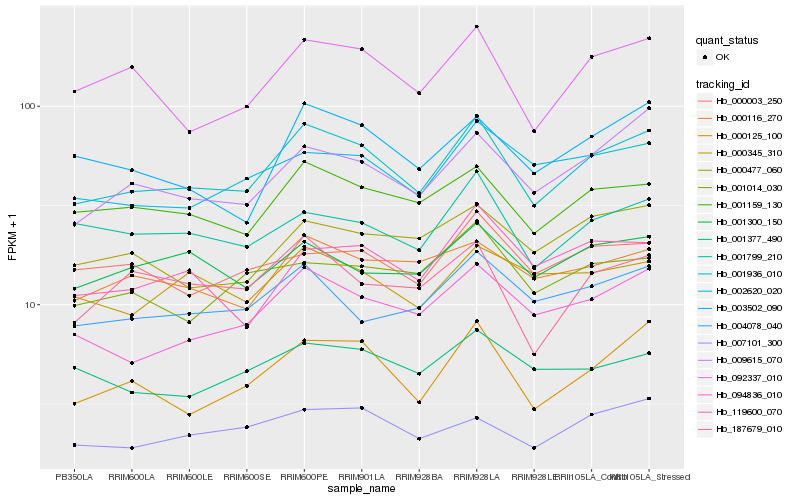
Hb_001936_010 |
0.0 |
- |
- |
hypothetical protein Csa_2G348280 [Cucumis sativus] |
| 2 |
Hb_001159_130 |
0.0660945552 |
- |
- |
PREDICTED: protein cornichon homolog 4 [Jatropha curcas] |
| 3 |
Hb_092337_010 |
0.0762800183 |
- |
- |
PREDICTED: actin-depolymerizing factor 2-like [Nelumbo nucifera] |
| 4 |
Hb_094836_010 |
0.0769400673 |
- |
- |
PREDICTED: uncharacterized protein LOC105638026 [Jatropha curcas] |
| 5 |
Hb_000116_270 |
0.0849470323 |
- |
- |
PREDICTED: SRSF protein kinase 2-like [Jatropha curcas] |
| 6 |
Hb_001799_210 |
0.0856742037 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 7 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002620_020 |
0.0873831923 |
transcription factor |
TF Family: bZIP |
DNA binding protein, putative [Ricinus communis] |
| 8 |
Hb_001377_490 |
0.0884027105 |
- |
- |
PREDICTED: uncharacterized protein LOC105642821 [Jatropha curcas] |
| 9 |
Hb_000345_310 |
0.0884652952 |
- |
- |
PREDICTED: sucrose nonfermenting 4-like protein isoform X3 [Jatropha curcas] |
| 10 |
Hb_000125_100 |
0.0889184214 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105635070 [Jatropha curcas] |
| 11 |
Hb_187679_010 |
0.0899173473 |
- |
- |
- |
| 12 |
Hb_000003_250 |
0.0903188517 |
- |
- |
glycerol-3-phosphate dehydrogenase, putative [Ricinus communis] |
| 13 |
Hb_004078_040 |
0.0903415887 |
- |
- |
PREDICTED: alkylated DNA repair protein alkB homolog 8 [Jatropha curcas] |
| 14 |
Hb_001014_030 |
0.0904669704 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 15 |
Hb_007101_300 |
0.0907564103 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g03880, mitochondrial [Jatropha curcas] |
| 16 |
Hb_003502_090 |
0.0918669189 |
- |
- |
ATP synthase subunit d, putative [Ricinus communis] |
| 17 |
Hb_119600_070 |
0.09195326 |
- |
- |
PREDICTED: uncharacterized protein LOC105635190 [Jatropha curcas] |
| 18 |
Hb_001300_150 |
0.0926771439 |
transcription factor |
TF Family: TUB |
PREDICTED: tubby-like F-box protein 3 [Jatropha curcas] |
| 19 |
Hb_000477_060 |
0.0935061945 |
- |
- |
PREDICTED: serine-threonine kinase receptor-associated protein-like [Jatropha curcas] |
| 20 |
Hb_009615_070 |
0.0941712925 |
- |
- |
type X protein phosphatase-II [Populus trichocarpa] |