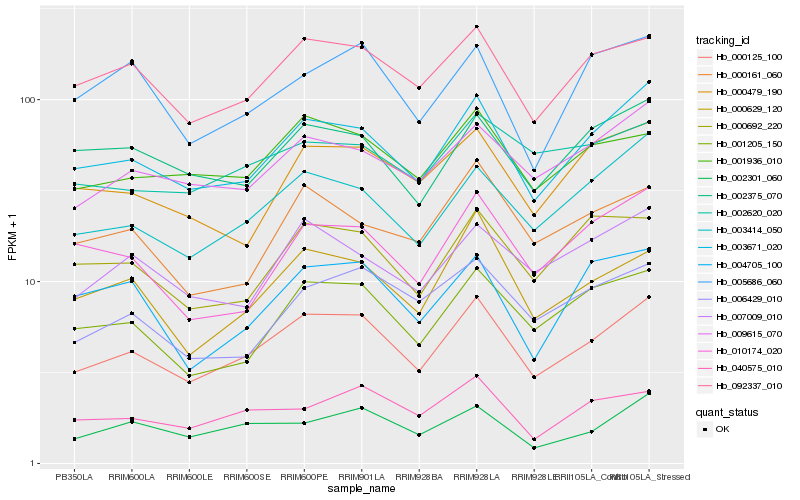
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000125_100 |
0.0 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105635070 [Jatropha curcas] |
| 2 |
Hb_003671_020 |
0.0568488769 |
- |
- |
calcineurin B, putative [Ricinus communis] |
| 3 |
Hb_092337_010 |
0.0787870882 |
- |
- |
PREDICTED: actin-depolymerizing factor 2-like [Nelumbo nucifera] |
| 4 |
Hb_003414_050 |
0.0796698169 |
- |
- |
PREDICTED: uncharacterized protein LOC105640180 isoform X3 [Jatropha curcas] |
| 5 |
Hb_001205_150 |
0.0879006836 |
- |
- |
Uncharacterized protein TCM_000403 [Theobroma cacao] |
| 6 |
Hb_009615_070 |
0.0881226115 |
- |
- |
type X protein phosphatase-II [Populus trichocarpa] |
| 7 |
Hb_000629_120 |
0.0883458196 |
- |
- |
Polynucleotidyl transferase, ribonuclease H-like superfamily protein [Theobroma cacao] |
| 8 |
Hb_001936_010 |
0.0889184214 |
- |
- |
hypothetical protein Csa_2G348280 [Cucumis sativus] |
| 9 |
Hb_002301_060 |
0.0951412557 |
- |
- |
hypothetical protein JCGZ_21483 [Jatropha curcas] |
| 10 |
Hb_040575_010 |
0.0989671838 |
- |
- |
PREDICTED: uncharacterized protein LOC105637675 [Jatropha curcas] |
| 11 |
Hb_002375_070 |
0.0997153717 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_007009_010 |
0.1003890075 |
transcription factor |
TF Family: TRAF |
PREDICTED: BTB/POZ domain-containing protein At3g05675 [Populus euphratica] |
| 13 |
Hb_006429_010 |
0.1014749454 |
- |
- |
PREDICTED: uncharacterized protein LOC105639724 [Jatropha curcas] |
| 14 |
Hb_000692_220 |
0.1016895994 |
- |
- |
hypothetical protein B456_002G120900 [Gossypium raimondii] |
| 15 |
Hb_000479_190 |
0.1025605556 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_010174_020 |
0.1071914727 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 17 |
Hb_000161_060 |
0.1081585349 |
- |
- |
PREDICTED: probable magnesium transporter NIPA4 [Jatropha curcas] |
| 18 |
Hb_005686_060 |
0.1084723439 |
- |
- |
PREDICTED: uncharacterized protein LOC101508199 [Cicer arietinum] |
| 19 |
Hb_002620_020 |
0.1085693927 |
transcription factor |
TF Family: bZIP |
DNA binding protein, putative [Ricinus communis] |
| 20 |
Hb_004705_100 |
0.1086303655 |
- |
- |
PREDICTED: inactive leucine-rich repeat receptor-like protein kinase CORYNE [Jatropha curcas] |