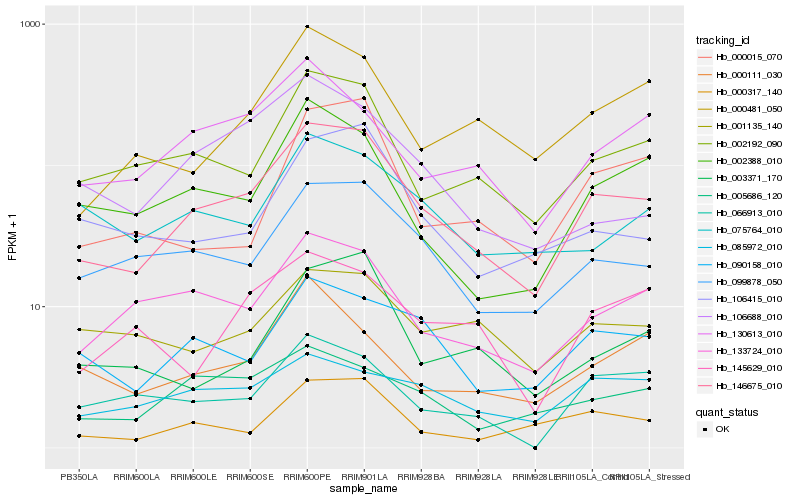
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_146675_010 |
0.0 |
- |
- |
PREDICTED: peroxisomal nicotinamide adenine dinucleotide carrier-like isoform X2 [Malus domestica] |
| 2 |
Hb_099878_050 |
0.1376039955 |
- |
- |
PREDICTED: uncharacterized protein LOC105123495 isoform X2 [Populus euphratica] |
| 3 |
Hb_002192_090 |
0.1383461644 |
- |
- |
hypothetical protein AMTR_s00029p00055000 [Amborella trichopoda] |
| 4 |
Hb_090158_010 |
0.1447256591 |
- |
- |
hypothetical protein RCOM_1968400 [Ricinus communis] |
| 5 |
Hb_133724_010 |
0.1478426481 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000317_140 |
0.1573760244 |
- |
- |
hypothetical protein CISIN_1g0019781mg, partial [Citrus sinensis] |
| 7 |
Hb_002388_010 |
0.160527443 |
- |
- |
- |
| 8 |
Hb_005686_120 |
0.1613067257 |
- |
- |
- |
| 9 |
Hb_000481_050 |
0.1630252616 |
- |
- |
PREDICTED: uncharacterized protein LOC105636546 [Jatropha curcas] |
| 10 |
Hb_106415_010 |
0.1645007677 |
- |
- |
PREDICTED: snRNA-activating protein complex subunit isoform X1 [Jatropha curcas] |
| 11 |
Hb_130613_010 |
0.1683545163 |
- |
- |
Charged multivesicular body protein 2a, putative [Ricinus communis] |
| 12 |
Hb_075764_010 |
0.1707823695 |
- |
- |
PREDICTED: PRA1 family protein A1-like [Jatropha curcas] |
| 13 |
Hb_003371_170 |
0.1727701556 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000015_070 |
0.1730521483 |
- |
- |
hypothetical protein POPTR_0001s22690g [Populus trichocarpa] |
| 15 |
Hb_085972_010 |
0.1735508889 |
transcription factor |
TF Family: NAC |
NAC transcription factor 066 [Jatropha curcas] |
| 16 |
Hb_066913_010 |
0.1782290935 |
- |
- |
hypothetical protein POPTR_0006s03150g [Populus trichocarpa] |
| 17 |
Hb_001135_140 |
0.1785145463 |
- |
- |
PREDICTED: serine/threonine-protein kinase PBS1-like [Malus domestica] |
| 18 |
Hb_145629_010 |
0.1801910659 |
- |
- |
- |
| 19 |
Hb_000111_030 |
0.1836478765 |
- |
- |
PREDICTED: uncharacterized protein LOC104094030 [Nicotiana tomentosiformis] |
| 20 |
Hb_106688_010 |
0.1837539009 |
- |
- |
aspartyl-tRNA synthetase, putative [Ricinus communis] |