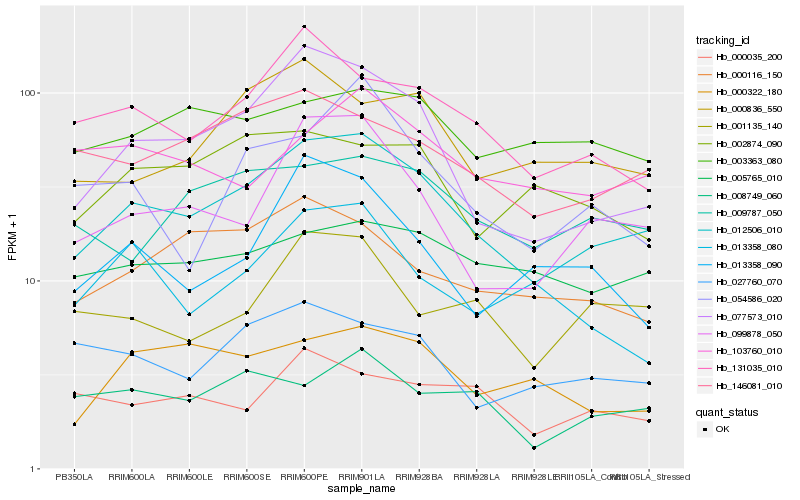
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012506_010 |
0.0 |
- |
- |
PREDICTED: UDP-D-apiose/UDP-D-xylose synthase 2 [Jatropha curcas] |
| 2 |
Hb_077573_010 |
0.1132964895 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 3 |
Hb_099878_050 |
0.1235494221 |
- |
- |
PREDICTED: uncharacterized protein LOC105123495 isoform X2 [Populus euphratica] |
| 4 |
Hb_131035_010 |
0.1293057444 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 5 |
Hb_146081_010 |
0.1309826826 |
- |
- |
PREDICTED: protein SCAI [Jatropha curcas] |
| 6 |
Hb_009787_050 |
0.1377639629 |
- |
- |
(Di)nucleoside polyphosphate hydrolase, putative [Ricinus communis] |
| 7 |
Hb_000836_550 |
0.139611056 |
- |
- |
PREDICTED: reticulon-like protein B5 [Jatropha curcas] |
| 8 |
Hb_000116_150 |
0.1400462923 |
- |
- |
serine/threonine-protein kinase cx32, putative [Ricinus communis] |
| 9 |
Hb_027760_070 |
0.1430720672 |
- |
- |
PREDICTED: uncharacterized endoplasmic reticulum membrane protein C16E8.02-like [Jatropha curcas] |
| 10 |
Hb_001135_140 |
0.1431589289 |
- |
- |
PREDICTED: serine/threonine-protein kinase PBS1-like [Malus domestica] |
| 11 |
Hb_005765_010 |
0.144378467 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At1g63170 isoform X1 [Jatropha curcas] |
| 12 |
Hb_008749_060 |
0.1475197971 |
- |
- |
PREDICTED: uncharacterized protein LOC105639821 [Jatropha curcas] |
| 13 |
Hb_000322_180 |
0.1479057523 |
- |
- |
recA family protein [Populus trichocarpa] |
| 14 |
Hb_002874_090 |
0.1504243427 |
- |
- |
PREDICTED: uncharacterized protein LOC105637452 [Jatropha curcas] |
| 15 |
Hb_003363_080 |
0.1523035136 |
- |
- |
eukaryotic translation elongation factor 1B gamma-subunit [Hevea brasiliensis] |
| 16 |
Hb_103760_010 |
0.1526742673 |
- |
- |
serine/arginine rich splicing factor, putative [Ricinus communis] |
| 17 |
Hb_013358_080 |
0.1533637283 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 25-like [Jatropha curcas] |
| 18 |
Hb_054586_020 |
0.1536989502 |
- |
- |
PREDICTED: exocyst complex component EXO70A1 [Jatropha curcas] |
| 19 |
Hb_013358_090 |
0.1546420282 |
- |
- |
hypothetical protein Csa_4G652025 [Cucumis sativus] |
| 20 |
Hb_000035_200 |
0.1553630611 |
- |
- |
PREDICTED: phospholipase D Z-like [Jatropha curcas] |