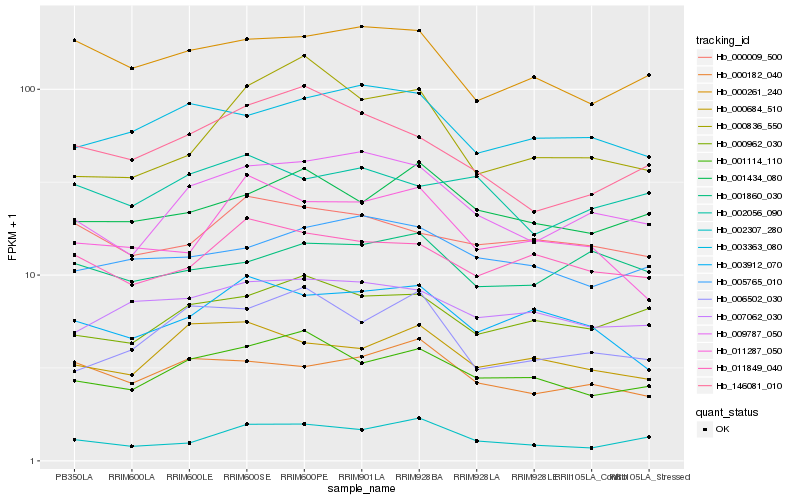
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009787_050 |
0.0 |
- |
- |
(Di)nucleoside polyphosphate hydrolase, putative [Ricinus communis] |
| 2 |
Hb_003363_080 |
0.096394489 |
- |
- |
eukaryotic translation elongation factor 1B gamma-subunit [Hevea brasiliensis] |
| 3 |
Hb_000962_030 |
0.0978815681 |
- |
- |
PREDICTED: metal tolerance protein C2 [Jatropha curcas] |
| 4 |
Hb_001114_110 |
0.1022553876 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2A isoform X1 [Jatropha curcas] |
| 5 |
Hb_005765_010 |
0.1077077558 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At1g63170 isoform X1 [Jatropha curcas] |
| 6 |
Hb_146081_010 |
0.1095919745 |
- |
- |
PREDICTED: protein SCAI [Jatropha curcas] |
| 7 |
Hb_002307_280 |
0.1118024587 |
- |
- |
Endonuclease/exonuclease/phosphatase family protein isoform 3 [Theobroma cacao] |
| 8 |
Hb_007062_030 |
0.1145214419 |
- |
- |
hypothetical protein VITISV_039033 [Vitis vinifera] |
| 9 |
Hb_003912_070 |
0.114627528 |
- |
- |
PREDICTED: nuclear pore complex protein NUP85 [Jatropha curcas] |
| 10 |
Hb_000182_040 |
0.1153357954 |
transcription factor |
TF Family: SNF2 |
PREDICTED: protein CHROMATIN REMODELING 24 [Jatropha curcas] |
| 11 |
Hb_006502_030 |
0.1162689172 |
- |
- |
PREDICTED: probable kinetochore protein ndc80 [Jatropha curcas] |
| 12 |
Hb_011849_040 |
0.117317985 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 13 |
Hb_000684_510 |
0.1196817783 |
- |
- |
run and tbc1 domain containing 3, plant, putative [Ricinus communis] |
| 14 |
Hb_001860_030 |
0.1200355963 |
- |
- |
PREDICTED: cleavage stimulation factor subunit 77 isoform X5 [Jatropha curcas] |
| 15 |
Hb_000261_240 |
0.1204529957 |
- |
- |
RecName: Full=Elongation factor 1-alpha; Short=EF-1-alpha [Manihot esculenta] |
| 16 |
Hb_000836_550 |
0.1227739885 |
- |
- |
PREDICTED: reticulon-like protein B5 [Jatropha curcas] |
| 17 |
Hb_001434_080 |
0.1230135752 |
- |
- |
PREDICTED: probable protein S-acyltransferase 7 [Jatropha curcas] |
| 18 |
Hb_000009_500 |
0.1230919248 |
- |
- |
PREDICTED: NASP-related protein sim3 [Jatropha curcas] |
| 19 |
Hb_002056_090 |
0.1232544599 |
transcription factor |
TF Family: GNAT |
PREDICTED: probable acetyltransferase NATA1-like [Jatropha curcas] |
| 20 |
Hb_011287_050 |
0.1236470782 |
- |
- |
PREDICTED: uncharacterized protein LOC105644786 [Jatropha curcas] |