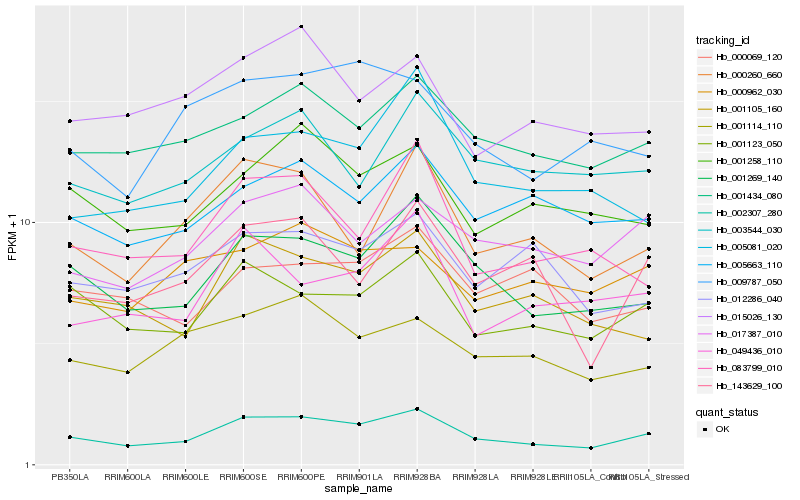
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002307_280 |
0.0 |
- |
- |
Endonuclease/exonuclease/phosphatase family protein isoform 3 [Theobroma cacao] |
| 2 |
Hb_001269_140 |
0.0774101895 |
- |
- |
PREDICTED: nuclear pore complex protein NUP96 [Jatropha curcas] |
| 3 |
Hb_001123_050 |
0.0904126928 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001434_080 |
0.0948091925 |
- |
- |
PREDICTED: probable protein S-acyltransferase 7 [Jatropha curcas] |
| 5 |
Hb_001105_160 |
0.1014786842 |
- |
- |
PREDICTED: protein phosphatase 2C 32 [Jatropha curcas] |
| 6 |
Hb_017387_010 |
0.1015790115 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At4g35230 [Jatropha curcas] |
| 7 |
Hb_001258_110 |
0.105804043 |
- |
- |
transporter, putative [Ricinus communis] |
| 8 |
Hb_005663_110 |
0.106478808 |
- |
- |
PREDICTED: large proline-rich protein BAG6 isoform X2 [Jatropha curcas] |
| 9 |
Hb_001114_110 |
0.1072231621 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2A isoform X1 [Jatropha curcas] |
| 10 |
Hb_000260_660 |
0.1080461292 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_143629_100 |
0.1095502564 |
- |
- |
PREDICTED: phytochrome A [Jatropha curcas] |
| 12 |
Hb_009787_050 |
0.1118024587 |
- |
- |
(Di)nucleoside polyphosphate hydrolase, putative [Ricinus communis] |
| 13 |
Hb_083799_010 |
0.1132889679 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 14 |
Hb_003544_030 |
0.1136475363 |
- |
- |
PREDICTED: citrate synthase, glyoxysomal [Jatropha curcas] |
| 15 |
Hb_005081_020 |
0.1139352559 |
- |
- |
PREDICTED: ninja-family protein mc410 [Jatropha curcas] |
| 16 |
Hb_012286_040 |
0.1151598514 |
- |
- |
PREDICTED: pre-mRNA-splicing factor RSE1 [Jatropha curcas] |
| 17 |
Hb_000962_030 |
0.1157243265 |
- |
- |
PREDICTED: metal tolerance protein C2 [Jatropha curcas] |
| 18 |
Hb_049436_010 |
0.1157708693 |
- |
- |
PREDICTED: UPF0202 protein At1g10490-like [Glycine max] |
| 19 |
Hb_015026_130 |
0.1159269233 |
- |
- |
quinone oxidoreductase, putative [Ricinus communis] |
| 20 |
Hb_000069_120 |
0.1165152584 |
- |
- |
PREDICTED: uncharacterized protein LOC105124742 isoform X1 [Populus euphratica] |