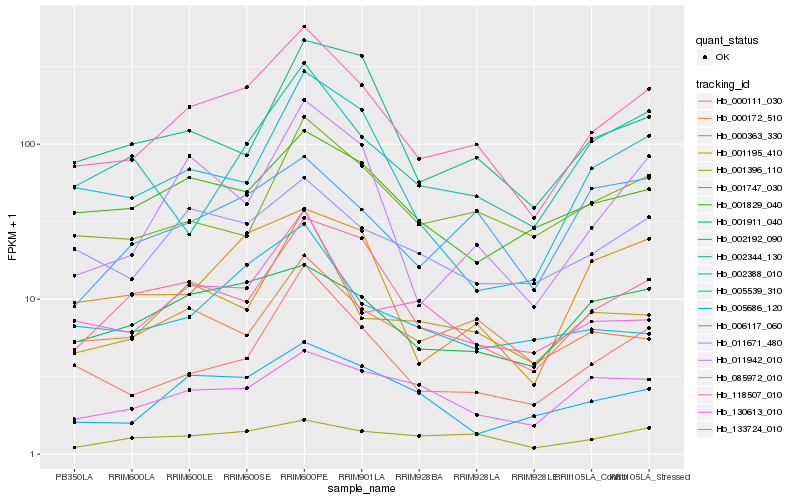
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_130613_010 |
0.0 |
- |
- |
Charged multivesicular body protein 2a, putative [Ricinus communis] |
| 2 |
Hb_000111_030 |
0.1225136187 |
- |
- |
PREDICTED: uncharacterized protein LOC104094030 [Nicotiana tomentosiformis] |
| 3 |
Hb_133724_010 |
0.1341793688 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_011671_480 |
0.1423429394 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000363_330 |
0.1426035336 |
- |
- |
- |
| 6 |
Hb_001911_040 |
0.1427971457 |
- |
- |
PREDICTED: uncharacterized protein LOC105628407 [Jatropha curcas] |
| 7 |
Hb_005686_120 |
0.1462664111 |
- |
- |
- |
| 8 |
Hb_011942_010 |
0.1462749238 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001195_410 |
0.1505012826 |
- |
- |
PREDICTED: deoxyuridine 5'-triphosphate nucleotidohydrolase [Jatropha curcas] |
| 10 |
Hb_001747_030 |
0.1515862728 |
- |
- |
hypothetical protein F383_08210 [Gossypium arboreum] |
| 11 |
Hb_001829_040 |
0.1519815449 |
- |
- |
PREDICTED: protein transport protein SEC13 homolog B [Jatropha curcas] |
| 12 |
Hb_002388_010 |
0.1569631697 |
- |
- |
- |
| 13 |
Hb_005539_310 |
0.1572534254 |
- |
- |
basic helix-loop-helix-containing protein, putative [Ricinus communis] |
| 14 |
Hb_118507_010 |
0.1589023407 |
- |
- |
hypothetical protein VITISV_019292 [Vitis vinifera] |
| 15 |
Hb_006117_060 |
0.1591444781 |
- |
- |
hypothetical protein CARUB_v10024452mg [Capsella rubella] |
| 16 |
Hb_002192_090 |
0.1594881657 |
- |
- |
hypothetical protein AMTR_s00029p00055000 [Amborella trichopoda] |
| 17 |
Hb_085972_010 |
0.159786308 |
transcription factor |
TF Family: NAC |
NAC transcription factor 066 [Jatropha curcas] |
| 18 |
Hb_001396_110 |
0.1629430585 |
- |
- |
DNA primase large subunit, putative [Ricinus communis] |
| 19 |
Hb_000172_510 |
0.1642485835 |
- |
- |
hypothetical protein B456_007G243100 [Gossypium raimondii] |
| 20 |
Hb_002344_130 |
0.1644697822 |
- |
- |
PREDICTED: copper transporter 5 [Jatropha curcas] |