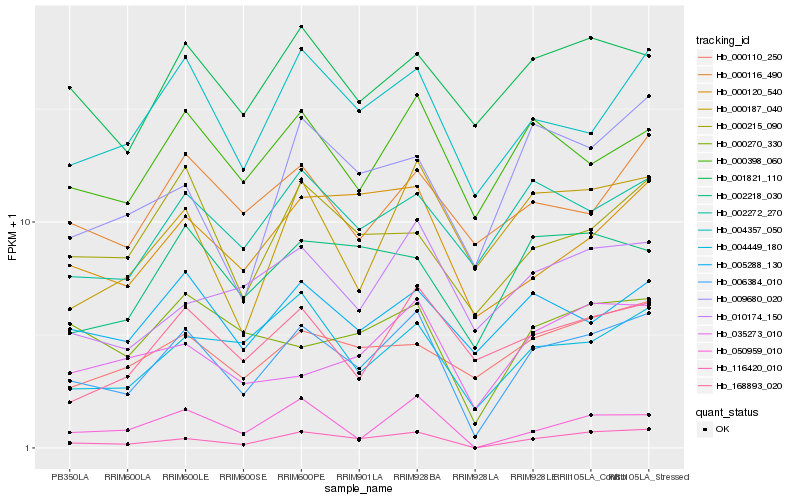
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006384_010 |
0.0 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 2 |
Hb_116420_010 |
0.1039100605 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |
| 3 |
Hb_004357_050 |
0.1351025475 |
- |
- |
PREDICTED: histone deacetylase HDT1-like [Jatropha curcas] |
| 4 |
Hb_004449_180 |
0.1422847535 |
- |
- |
PREDICTED: SAC3 family protein 1 [Jatropha curcas] |
| 5 |
Hb_000398_060 |
0.1460853248 |
- |
- |
PREDICTED: survival of motor neuron-related-splicing factor 30 [Jatropha curcas] |
| 6 |
Hb_002272_270 |
0.1527957134 |
- |
- |
hypothetical protein POPTR_0013s02780g [Populus trichocarpa] |
| 7 |
Hb_005288_130 |
0.1528385354 |
- |
- |
protein with unknown function [Ricinus communis] |
| 8 |
Hb_000270_330 |
0.153937292 |
- |
- |
ribosomal protein S6 kinase, putative [Ricinus communis] |
| 9 |
Hb_050959_010 |
0.1543137007 |
- |
- |
- |
| 10 |
Hb_000120_540 |
0.1545815467 |
- |
- |
translation initiation factor, putative [Ricinus communis] |
| 11 |
Hb_000187_040 |
0.1547267519 |
- |
- |
PREDICTED: pyridoxal kinase isoform X5 [Vitis vinifera] |
| 12 |
Hb_000215_090 |
0.1565698436 |
- |
- |
cytochrome b5 domain-containing family protein [Populus trichocarpa] |
| 13 |
Hb_002218_030 |
0.1578139065 |
- |
- |
hypothetical protein B456_007G345200 [Gossypium raimondii] |
| 14 |
Hb_001821_110 |
0.1595976587 |
- |
- |
PREDICTED: ubiquinol oxidase, mitochondrial [Jatropha curcas] |
| 15 |
Hb_168893_020 |
0.1603120726 |
- |
- |
PREDICTED: probable protein S-acyltransferase 15 [Jatropha curcas] |
| 16 |
Hb_035273_010 |
0.1608339827 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647563 [Jatropha curcas] |
| 17 |
Hb_010174_150 |
0.1614015537 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 18 |
Hb_009680_020 |
0.1617616781 |
- |
- |
PREDICTED: 40S ribosomal protein S10 [Jatropha curcas] |
| 19 |
Hb_000116_490 |
0.1641374081 |
- |
- |
PREDICTED: WD repeat-containing protein 55 homolog [Jatropha curcas] |
| 20 |
Hb_000110_250 |
0.1646755191 |
- |
- |
PREDICTED: homologous-pairing protein 2 homolog [Jatropha curcas] |