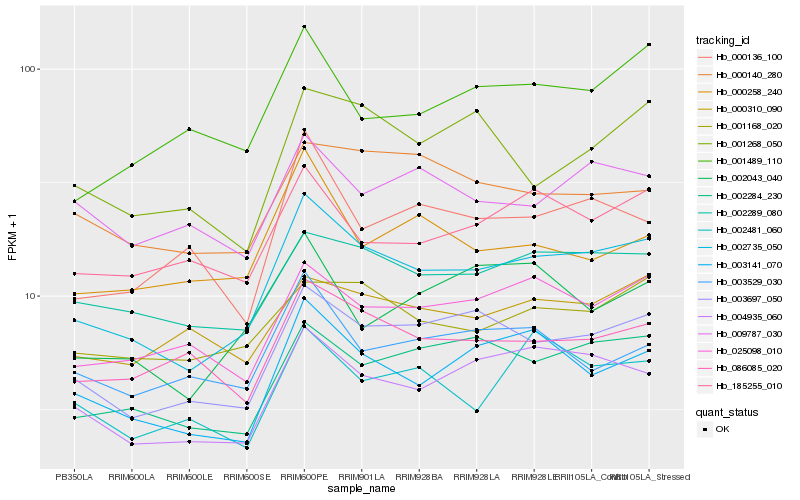
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002735_050 |
0.0 |
- |
- |
PREDICTED: thiamine-repressible mitochondrial transport protein THI74 isoform X1 [Jatropha curcas] |
| 2 |
Hb_003697_050 |
0.0823667023 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 13 [Jatropha curcas] |
| 3 |
Hb_004935_060 |
0.0880679275 |
- |
- |
Uncharacterized protein isoform 2 [Theobroma cacao] |
| 4 |
Hb_003141_070 |
0.0902661336 |
- |
- |
PREDICTED: probable protein phosphatase 2C 51 [Jatropha curcas] |
| 5 |
Hb_002289_080 |
0.0934892375 |
- |
- |
hypothetical protein POPTR_0016s05590g [Populus trichocarpa] |
| 6 |
Hb_002284_230 |
0.095634663 |
- |
- |
PREDICTED: uncharacterized protein At5g43822 [Jatropha curcas] |
| 7 |
Hb_025098_010 |
0.1030367631 |
- |
- |
PREDICTED: protoheme IX farnesyltransferase, mitochondrial isoform X2 [Jatropha curcas] |
| 8 |
Hb_086085_020 |
0.1088642164 |
- |
- |
Non-imprinted in Prader-Willi/Angelman syndrome region protein isoform 3 [Theobroma cacao] |
| 9 |
Hb_001168_020 |
0.1097613248 |
- |
- |
PREDICTED: translation initiation factor eIF-2B subunit gamma [Jatropha curcas] |
| 10 |
Hb_185255_010 |
0.1104498666 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 11-like [Jatropha curcas] |
| 11 |
Hb_000140_280 |
0.1165685221 |
- |
- |
hypothetical protein B456_001G157900 [Gossypium raimondii] |
| 12 |
Hb_001268_050 |
0.1174794758 |
- |
- |
translocon-associated protein, beta subunit precursor, putative [Ricinus communis] |
| 13 |
Hb_001489_110 |
0.1180691129 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin of 36 kDa-like [Citrus sinensis] |
| 14 |
Hb_002043_040 |
0.1184551432 |
- |
- |
PREDICTED: uncharacterized protein LOC105631509 [Jatropha curcas] |
| 15 |
Hb_000310_090 |
0.1189520457 |
- |
- |
3-oxoacyl-[acyl-carrier-protein] synthase 3 A, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000136_100 |
0.120352861 |
- |
- |
PREDICTED: syntaxin-61 [Jatropha curcas] |
| 17 |
Hb_003529_030 |
0.1204118698 |
- |
- |
Non-imprinted in Prader-Willi/Angelman syndrome region protein, putative [Ricinus communis] |
| 18 |
Hb_002481_060 |
0.1211354868 |
- |
- |
PREDICTED: uncharacterized protein LOC105640851 [Jatropha curcas] |
| 19 |
Hb_000258_240 |
0.1216808241 |
- |
- |
sur2 hydroxylase/desaturase, putative [Ricinus communis] |
| 20 |
Hb_009787_030 |
0.1228717354 |
- |
- |
Prolactin regulatory element-binding protein, putative [Ricinus communis] |