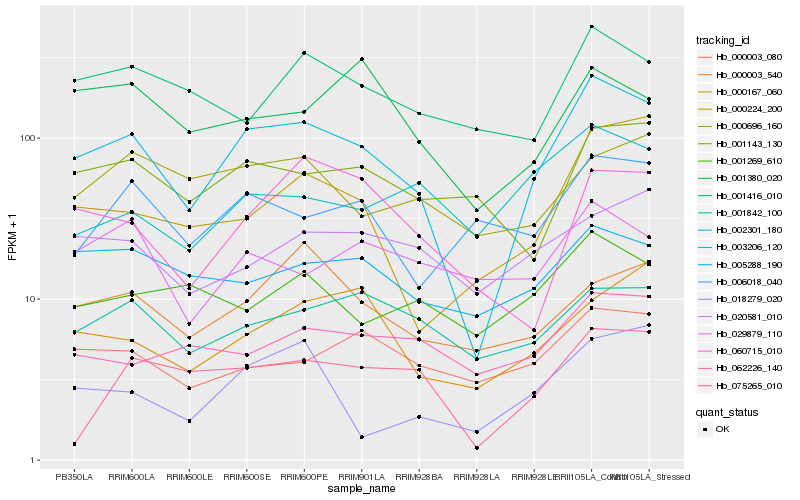
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003206_120 |
0.0 |
- |
- |
PREDICTED: glutamate decarboxylase 5-like [Jatropha curcas] |
| 2 |
Hb_000224_200 |
0.1845687013 |
- |
- |
PREDICTED: uncharacterized protein LOC105640874 [Jatropha curcas] |
| 3 |
Hb_000003_080 |
0.1865119666 |
- |
- |
PREDICTED: SART-1 family protein DOT2 isoform X1 [Jatropha curcas] |
| 4 |
Hb_005288_190 |
0.1876254104 |
- |
- |
PREDICTED: exportin-4 [Jatropha curcas] |
| 5 |
Hb_001842_100 |
0.1906619573 |
rubber biosynthesis |
Gene Name: Dimethyallyltransferase |
hypothetical protein JCGZ_10905 [Jatropha curcas] |
| 6 |
Hb_001416_010 |
0.1912617902 |
- |
- |
Thioredoxin H-type, putative [Ricinus communis] |
| 7 |
Hb_001143_130 |
0.193506302 |
- |
- |
peroxidase 2 [Hevea brasiliensis] |
| 8 |
Hb_000167_060 |
0.1937401742 |
- |
- |
PREDICTED: U11/U12 small nuclear ribonucleoprotein 31 kDa protein [Jatropha curcas] |
| 9 |
Hb_060715_010 |
0.1952067016 |
- |
- |
PREDICTED: notchless protein homolog [Gossypium raimondii] |
| 10 |
Hb_075265_010 |
0.1955316255 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 11 |
Hb_018279_020 |
0.1971862738 |
- |
- |
hypothetical protein JCGZ_03682 [Jatropha curcas] |
| 12 |
Hb_000003_540 |
0.1972883768 |
- |
- |
PREDICTED: homeobox-leucine zipper protein ATHB-15 [Jatropha curcas] |
| 13 |
Hb_020581_010 |
0.1973635953 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 14 |
Hb_062226_140 |
0.1990466063 |
- |
- |
PREDICTED: protein NEDD1 [Jatropha curcas] |
| 15 |
Hb_001269_610 |
0.2004659651 |
- |
- |
Chaperone protein dnaJ 8, chloroplast precursor, putative [Ricinus communis] |
| 16 |
Hb_001380_020 |
0.2007858567 |
- |
- |
3-isopropylmalate dehydratase, putative [Ricinus communis] |
| 17 |
Hb_006018_040 |
0.2015058094 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase HIP1 [Jatropha curcas] |
| 18 |
Hb_002301_180 |
0.2038350704 |
- |
- |
PREDICTED: protein yippee-like At5g53940 [Jatropha curcas] |
| 19 |
Hb_029879_110 |
0.2038394005 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g61870, mitochondrial [Jatropha curcas] |
| 20 |
Hb_000696_160 |
0.2043979661 |
- |
- |
PREDICTED: SKP1-like protein 1B [Jatropha curcas] |