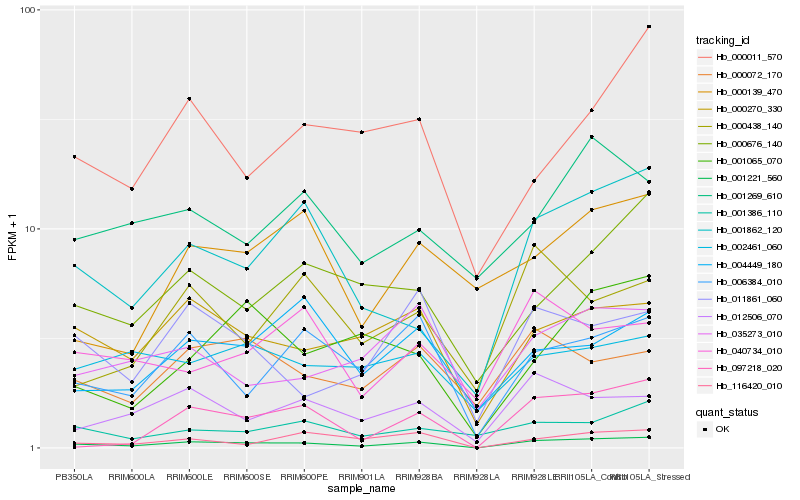
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001221_560 |
0.0 |
- |
- |
PREDICTED: 11-beta-hydroxysteroid dehydrogenase 1B-like [Jatropha curcas] |
| 2 |
Hb_001862_120 |
0.1687068484 |
- |
- |
PREDICTED: aspartic proteinase-like protein 2 [Jatropha curcas] |
| 3 |
Hb_000270_330 |
0.1706314285 |
- |
- |
ribosomal protein S6 kinase, putative [Ricinus communis] |
| 4 |
Hb_006384_010 |
0.1764044028 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 5 |
Hb_012506_070 |
0.188971906 |
- |
- |
hypothetical protein POPTR_0004s20060g [Populus trichocarpa] |
| 6 |
Hb_000676_140 |
0.1930609396 |
- |
- |
structural constituent of ribosome, putative [Ricinus communis] |
| 7 |
Hb_116420_010 |
0.1932904024 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |
| 8 |
Hb_011861_060 |
0.1941209066 |
- |
- |
- |
| 9 |
Hb_001386_110 |
0.1967212145 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: putative fasciclin-like arabinogalactan protein 20 [Populus euphratica] |
| 10 |
Hb_097218_020 |
0.199432354 |
- |
- |
PREDICTED: replication protein A 70 kDa DNA-binding subunit E [Jatropha curcas] |
| 11 |
Hb_035273_010 |
0.2006105834 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647563 [Jatropha curcas] |
| 12 |
Hb_000072_170 |
0.2006286934 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |
| 13 |
Hb_004449_180 |
0.2061212104 |
- |
- |
PREDICTED: SAC3 family protein 1 [Jatropha curcas] |
| 14 |
Hb_001065_070 |
0.2066590135 |
- |
- |
- |
| 15 |
Hb_000011_570 |
0.2088887408 |
- |
- |
PREDICTED: 40S ribosomal protein SA-like [Jatropha curcas] |
| 16 |
Hb_001269_610 |
0.2112232619 |
- |
- |
Chaperone protein dnaJ 8, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_000438_140 |
0.2125676039 |
- |
- |
hypothetical protein CISIN_1g018088mg [Citrus sinensis] |
| 18 |
Hb_040734_010 |
0.2131732892 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g37230 [Jatropha curcas] |
| 19 |
Hb_000139_470 |
0.2136092638 |
- |
- |
Dual specificity protein phosphatase, putative [Ricinus communis] |
| 20 |
Hb_002461_060 |
0.214415374 |
- |
- |
PREDICTED: SWR1-complex protein 4 isoform X3 [Jatropha curcas] |