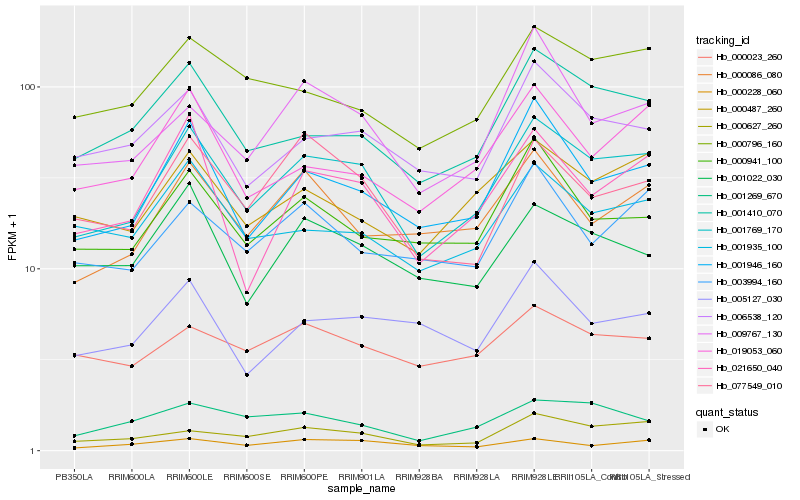
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001769_170 |
0.0 |
- |
- |
50S ribosomal protein L31, putative [Ricinus communis] |
| 2 |
Hb_001946_160 |
0.0955119559 |
- |
- |
putative chaperon P13.9 [Castanea sativa] |
| 3 |
Hb_001410_070 |
0.1018435878 |
- |
- |
hypothetical protein JCGZ_20421 [Jatropha curcas] |
| 4 |
Hb_000627_260 |
0.1067028871 |
- |
- |
chloride channel clc, putative [Ricinus communis] |
| 5 |
Hb_021650_040 |
0.1084910888 |
- |
- |
EG2771 [Manihot esculenta] |
| 6 |
Hb_000487_260 |
0.10861423 |
- |
- |
PREDICTED: protein Iojap, chloroplastic [Jatropha curcas] |
| 7 |
Hb_006538_120 |
0.1086737621 |
- |
- |
PREDICTED: 50S ribosomal protein L15, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000796_160 |
0.1112387142 |
- |
- |
PREDICTED: nifU-like protein 1, chloroplastic [Jatropha curcas] |
| 9 |
Hb_077549_010 |
0.1114085777 |
- |
- |
PREDICTED: uncharacterized protein LOC105644817 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001935_100 |
0.113753229 |
- |
- |
structural molecule, putative [Ricinus communis] |
| 11 |
Hb_000086_080 |
0.1141714941 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000941_100 |
0.1163150922 |
- |
- |
thioredoxin-like 5 mRNA family protein [Populus trichocarpa] |
| 13 |
Hb_019053_060 |
0.117277322 |
- |
- |
PREDICTED: uncharacterized protein LOC105632672 [Jatropha curcas] |
| 14 |
Hb_001022_030 |
0.1173393791 |
- |
- |
PREDICTED: sorting nexin 1 [Jatropha curcas] |
| 15 |
Hb_003994_160 |
0.1202627428 |
- |
- |
PREDICTED: uncharacterized protein LOC105131691 isoform X5 [Populus euphratica] |
| 16 |
Hb_000023_260 |
0.1215197653 |
- |
- |
PREDICTED: monogalactosyldiacylglycerol synthase, chloroplastic [Jatropha curcas] |
| 17 |
Hb_001269_670 |
0.122957428 |
- |
- |
PREDICTED: uncharacterized protein LOC105630287 [Jatropha curcas] |
| 18 |
Hb_009767_130 |
0.1243124003 |
- |
- |
PREDICTED: preprotein translocase subunit SECE1 [Jatropha curcas] |
| 19 |
Hb_000228_060 |
0.1250477898 |
- |
- |
hypothetical protein JCGZ_21532 [Jatropha curcas] |
| 20 |
Hb_005127_030 |
0.1259197369 |
- |
- |
PREDICTED: tryptophan--tRNA ligase, mitochondrial isoform X2 [Jatropha curcas] |