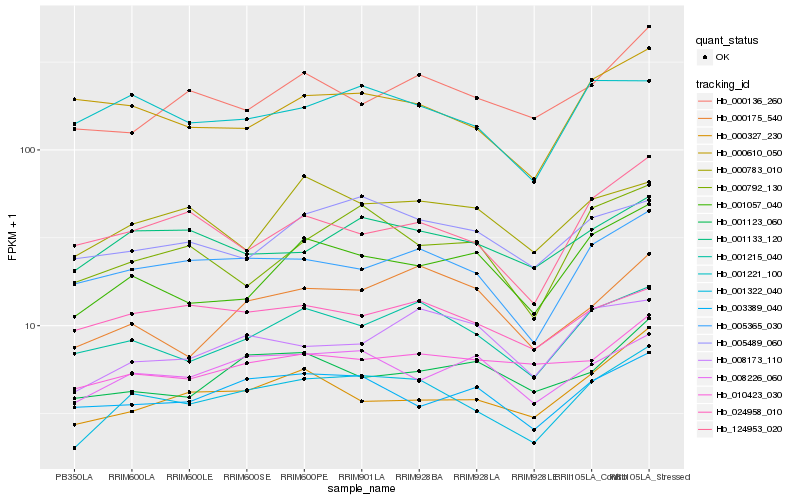
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001322_040 |
0.0 |
- |
- |
- |
| 2 |
Hb_001215_040 |
0.100376021 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_005365_030 |
0.1006183501 |
- |
- |
hypothetical protein POPTR_0002s24010g [Populus trichocarpa] |
| 4 |
Hb_008226_060 |
0.1119875504 |
- |
- |
PREDICTED: folic acid synthesis protein fol1-like [Jatropha curcas] |
| 5 |
Hb_001057_040 |
0.115447938 |
- |
- |
PREDICTED: oligosaccharyltransferase complex subunit ostc [Jatropha curcas] |
| 6 |
Hb_003389_040 |
0.1173283419 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 12 [Jatropha curcas] |
| 7 |
Hb_001133_120 |
0.1186784577 |
- |
- |
PREDICTED: uncharacterized protein LOC105629988 [Jatropha curcas] |
| 8 |
Hb_000175_540 |
0.1192521718 |
- |
- |
PREDICTED: uncharacterized protein LOC105634886 [Jatropha curcas] |
| 9 |
Hb_000327_230 |
0.1206902755 |
- |
- |
PREDICTED: nitrilase-like protein 2 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001221_100 |
0.1207453053 |
- |
- |
PREDICTED: elongation factor 1-delta 1-like [Citrus sinensis] |
| 11 |
Hb_000792_130 |
0.1214033221 |
- |
- |
60S ribosomal L35a-3 -like protein [Gossypium arboreum] |
| 12 |
Hb_005489_060 |
0.122653077 |
- |
- |
Anamorsin, putative [Ricinus communis] |
| 13 |
Hb_124953_020 |
0.1237527351 |
- |
- |
Mitochondrial import inner membrane translocase subunit Tim13, putative [Ricinus communis] |
| 14 |
Hb_008173_110 |
0.1242674105 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 15 |
Hb_000783_010 |
0.1278659005 |
- |
- |
PREDICTED: ras-related protein RABF1 [Jatropha curcas] |
| 16 |
Hb_000136_260 |
0.1282286574 |
- |
- |
40S ribosomal protein S5A [Hevea brasiliensis] |
| 17 |
Hb_024958_010 |
0.1288204421 |
- |
- |
PREDICTED: LRR repeats and ubiquitin-like domain-containing protein At2g30105 [Jatropha curcas] |
| 18 |
Hb_010423_030 |
0.1292881697 |
- |
- |
PREDICTED: uncharacterized protein LOC105645584 [Jatropha curcas] |
| 19 |
Hb_001123_060 |
0.1311952484 |
- |
- |
PREDICTED: ALBINO3-like protein 2, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000610_050 |
0.1313400434 |
- |
- |
skp1, putative [Ricinus communis] |