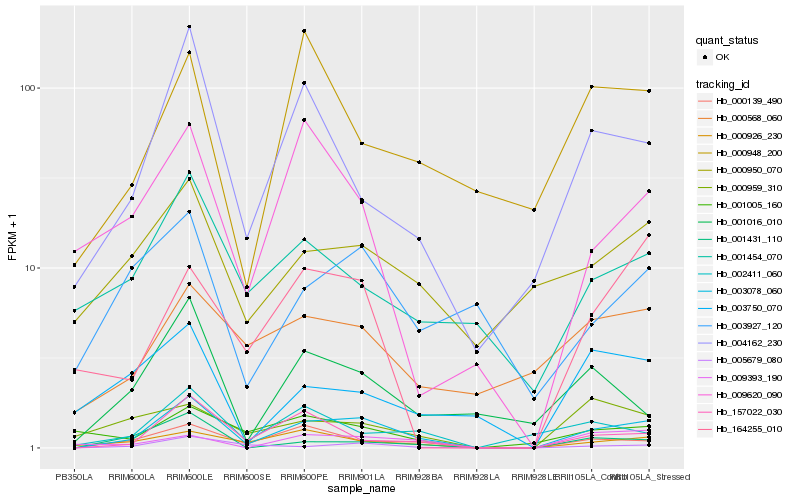
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003078_060 |
0.0 |
- |
- |
60S ribosomal protein L32, putative [Ricinus communis] |
| 2 |
Hb_000139_490 |
0.2192959841 |
- |
- |
PREDICTED: transcription factor MUTE [Jatropha curcas] |
| 3 |
Hb_157022_030 |
0.238149452 |
transcription factor |
TF Family: NF-YB |
ccaat-binding transcription factor subunit A, putative [Ricinus communis] |
| 4 |
Hb_004162_230 |
0.2424526984 |
- |
- |
PREDICTED: histone H2B [Jatropha curcas] |
| 5 |
Hb_000926_230 |
0.2464225391 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_005679_080 |
0.2487097974 |
- |
- |
PREDICTED: PH-interacting protein-like [Populus euphratica] |
| 7 |
Hb_001431_110 |
0.2625024234 |
- |
- |
glycerol-3-phosphate transporter family protein [Populus trichocarpa] |
| 8 |
Hb_002411_060 |
0.2724325376 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 5 [Jatropha curcas] |
| 9 |
Hb_009393_190 |
0.2755372359 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase AIP2-like [Jatropha curcas] |
| 10 |
Hb_001016_010 |
0.2757527289 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000568_060 |
0.2847404998 |
- |
- |
PREDICTED: uncharacterized protein LOC105650906 [Jatropha curcas] |
| 12 |
Hb_001454_070 |
0.284830983 |
- |
- |
Uveal autoantigen with coiled-coil domains and ankyrin repeats isoform 1 [Theobroma cacao] |
| 13 |
Hb_000950_070 |
0.2848799015 |
- |
- |
PREDICTED: uncharacterized protein LOC105645259 isoform X2 [Jatropha curcas] |
| 14 |
Hb_164255_010 |
0.2850659873 |
- |
- |
PREDICTED: aspartate-semialdehyde dehydrogenase [Jatropha curcas] |
| 15 |
Hb_001005_160 |
0.2860348226 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_003927_120 |
0.2866207147 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620 [Jatropha curcas] |
| 17 |
Hb_009620_090 |
0.2895877029 |
- |
- |
hypothetical protein L484_000980 [Morus notabilis] |
| 18 |
Hb_000948_200 |
0.291716338 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |
| 19 |
Hb_000959_310 |
0.2925382948 |
- |
- |
PREDICTED: WD repeat domain-containing protein 83 [Jatropha curcas] |
| 20 |
Hb_003750_070 |
0.2929605092 |
- |
- |
PREDICTED: psbQ-like protein 3, chloroplastic [Jatropha curcas] |