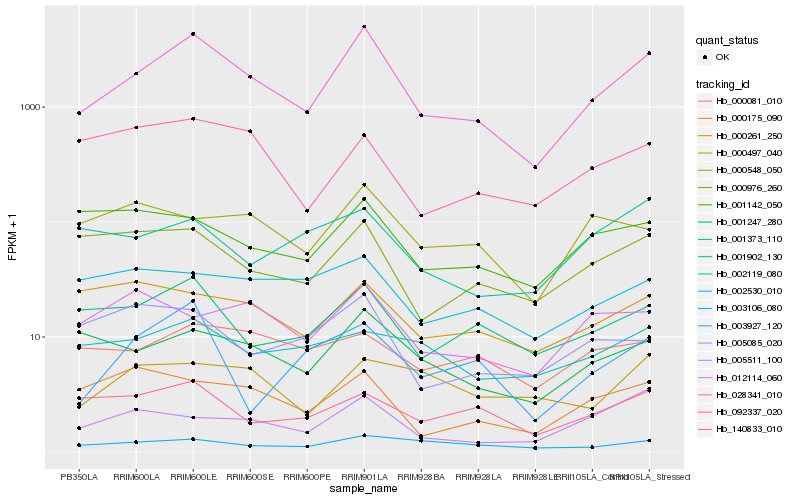
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_028341_010 |
0.0 |
- |
- |
hypothetical protein CICLE_v10026841mg [Citrus clementina] |
| 2 |
Hb_001247_280 |
0.168163048 |
- |
- |
PREDICTED: uncharacterized protein LOC105632071 [Jatropha curcas] |
| 3 |
Hb_001373_110 |
0.1710681399 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 8 [Jatropha curcas] |
| 4 |
Hb_000976_260 |
0.1857688119 |
- |
- |
- |
| 5 |
Hb_003106_080 |
0.1877519928 |
- |
- |
cell division cycle, putative [Ricinus communis] |
| 6 |
Hb_005511_100 |
0.1917738484 |
- |
- |
PREDICTED: uncharacterized protein LOC105645493 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000175_090 |
0.1960191672 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_140833_010 |
0.1977824132 |
- |
- |
PREDICTED: uncharacterized protein LOC105647767 [Jatropha curcas] |
| 9 |
Hb_002530_010 |
0.2007791374 |
- |
- |
hypothetical protein EUGRSUZ_C02580 [Eucalyptus grandis] |
| 10 |
Hb_012114_060 |
0.2018375645 |
- |
- |
PREDICTED: uncharacterized protein LOC105645814 [Jatropha curcas] |
| 11 |
Hb_000548_050 |
0.2020735137 |
- |
- |
hypothetical protein POPTR_0008s12570g [Populus trichocarpa] |
| 12 |
Hb_092337_020 |
0.2024893417 |
- |
- |
hypothetical protein B456_013G264900 [Gossypium raimondii] |
| 13 |
Hb_000261_250 |
0.2040205488 |
- |
- |
elongation factor 1-alpha [Lepidium sativum] |
| 14 |
Hb_000497_040 |
0.2044579183 |
- |
- |
BnaC09g09960D [Brassica napus] |
| 15 |
Hb_001142_050 |
0.2047672104 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 12 [Jatropha curcas] |
| 16 |
Hb_002119_080 |
0.2053324202 |
- |
- |
PREDICTED: 54S ribosomal protein L24, mitochondrial [Jatropha curcas] |
| 17 |
Hb_005085_020 |
0.2060664814 |
- |
- |
hypothetical protein B456_001G193200 [Gossypium raimondii] |
| 18 |
Hb_000081_010 |
0.2062678257 |
- |
- |
hypothetical protein B456_012G056000 [Gossypium raimondii] |
| 19 |
Hb_003927_120 |
0.2063449254 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620 [Jatropha curcas] |
| 20 |
Hb_001902_130 |
0.208197093 |
- |
- |
60S ribosomal protein L36-2 [Morus notabilis] |