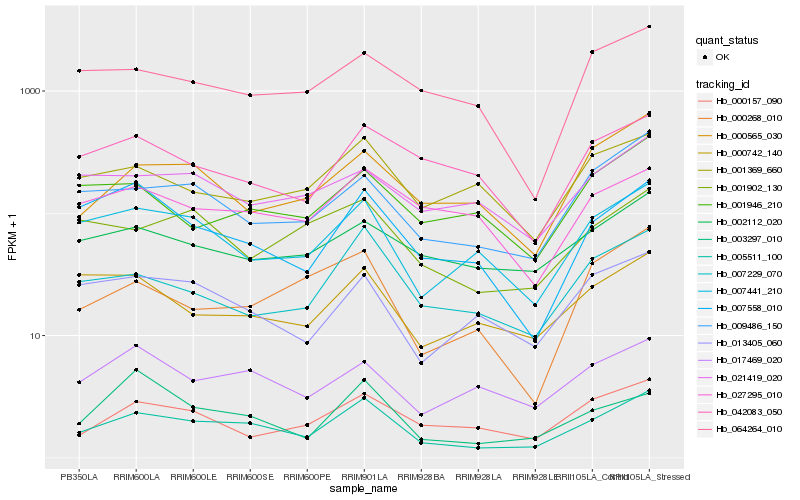
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005511_100 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105645493 isoform X1 [Jatropha curcas] |
| 2 |
Hb_007441_210 |
0.1364932157 |
- |
- |
hypothetical protein PRUPE_ppa013350mg [Prunus persica] |
| 3 |
Hb_009486_150 |
0.1366056263 |
- |
- |
60S ribosomal protein L37a-2 [Medicago truncatula] |
| 4 |
Hb_007229_070 |
0.1367037792 |
- |
- |
PREDICTED: UPF0565 protein C2orf69 [Jatropha curcas] |
| 5 |
Hb_000565_030 |
0.1510602563 |
- |
- |
PREDICTED: 40S ribosomal protein S26-3-like [Jatropha curcas] |
| 6 |
Hb_007558_010 |
0.1517147569 |
- |
- |
PREDICTED: mitochondrial import receptor subunit TOM9-2 [Jatropha curcas] |
| 7 |
Hb_017469_020 |
0.1530391257 |
- |
- |
- |
| 8 |
Hb_000157_090 |
0.1543875804 |
- |
- |
PREDICTED: 2-aminoethanethiol dioxygenase-like [Jatropha curcas] |
| 9 |
Hb_013405_060 |
0.1552495315 |
- |
- |
PREDICTED: probable RNA-binding protein 18 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000268_010 |
0.157575907 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000742_140 |
0.1589905832 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002112_020 |
0.1593988339 |
- |
- |
60S ribosomal protein L9, putative [Ricinus communis] |
| 13 |
Hb_001902_130 |
0.160678748 |
- |
- |
60S ribosomal protein L36-2 [Morus notabilis] |
| 14 |
Hb_001369_660 |
0.1622757086 |
- |
- |
hypothetical protein B456_001G174200, partial [Gossypium raimondii] |
| 15 |
Hb_064264_010 |
0.1627163642 |
- |
- |
hypothetical protein [Zea mays] |
| 16 |
Hb_001946_210 |
0.1654047982 |
- |
- |
ribosomal protein l7ae, putative [Ricinus communis] |
| 17 |
Hb_027295_010 |
0.1662257818 |
- |
- |
40S ribosomal protein S7, putative [Ricinus communis] |
| 18 |
Hb_042083_050 |
0.1662941269 |
- |
- |
60S ribosomal protein L11, putative [Ricinus communis] |
| 19 |
Hb_003297_010 |
0.1671285675 |
- |
- |
PREDICTED: uncharacterized protein LOC105636715 [Jatropha curcas] |
| 20 |
Hb_021419_020 |
0.1689602498 |
- |
- |
PREDICTED: 60S ribosomal protein L13-1-like [Jatropha curcas] |