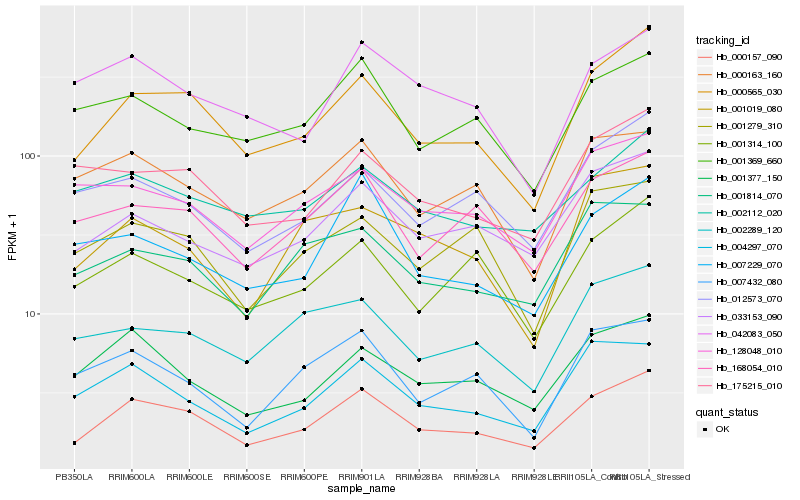
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000157_090 |
0.0 |
- |
- |
PREDICTED: 2-aminoethanethiol dioxygenase-like [Jatropha curcas] |
| 2 |
Hb_000565_030 |
0.0690919998 |
- |
- |
PREDICTED: 40S ribosomal protein S26-3-like [Jatropha curcas] |
| 3 |
Hb_033153_090 |
0.1081580083 |
- |
- |
PREDICTED: 28 kDa heat- and acid-stable phosphoprotein [Jatropha curcas] |
| 4 |
Hb_001377_150 |
0.1110511521 |
- |
- |
hypothetical protein JCGZ_14266 [Jatropha curcas] |
| 5 |
Hb_004297_070 |
0.1121077983 |
- |
- |
hypothetical protein POPTR_0009s14820g [Populus trichocarpa] |
| 6 |
Hb_168054_010 |
0.1139340148 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP18-1 [Elaeis guineensis] |
| 7 |
Hb_001314_100 |
0.1226914428 |
- |
- |
hypothetical protein POPTR_0001s01820g [Populus trichocarpa] |
| 8 |
Hb_012573_070 |
0.1254646209 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001369_660 |
0.1264945949 |
- |
- |
hypothetical protein B456_001G174200, partial [Gossypium raimondii] |
| 10 |
Hb_001279_310 |
0.1269074562 |
- |
- |
hypothetical protein JCGZ_10212 [Jatropha curcas] |
| 11 |
Hb_007432_080 |
0.1269835265 |
- |
- |
PREDICTED: mannose-6-phosphate isomerase 1-like [Jatropha curcas] |
| 12 |
Hb_000163_160 |
0.1273153837 |
- |
- |
Putative small nuclear ribonucleoprotein Sm D2 [Glycine soja] |
| 13 |
Hb_001814_070 |
0.1275178685 |
- |
- |
PREDICTED: uncharacterized protein LOC105650638 [Jatropha curcas] |
| 14 |
Hb_175215_010 |
0.1278057782 |
- |
- |
hypothetical protein CISIN_1g029605mg [Citrus sinensis] |
| 15 |
Hb_007229_070 |
0.1290390471 |
- |
- |
PREDICTED: UPF0565 protein C2orf69 [Jatropha curcas] |
| 16 |
Hb_002112_020 |
0.1290653301 |
- |
- |
60S ribosomal protein L9, putative [Ricinus communis] |
| 17 |
Hb_002289_120 |
0.1294952208 |
- |
- |
PREDICTED: uncharacterized protein LOC105645218 isoform X2 [Jatropha curcas] |
| 18 |
Hb_001019_080 |
0.1299643621 |
- |
- |
hypothetical protein JCGZ_13280 [Jatropha curcas] |
| 19 |
Hb_128048_010 |
0.1312498082 |
- |
- |
small nuclear ribonucleoprotein U)1a,U)2b, putative [Ricinus communis] |
| 20 |
Hb_042083_050 |
0.1323860408 |
- |
- |
60S ribosomal protein L11, putative [Ricinus communis] |