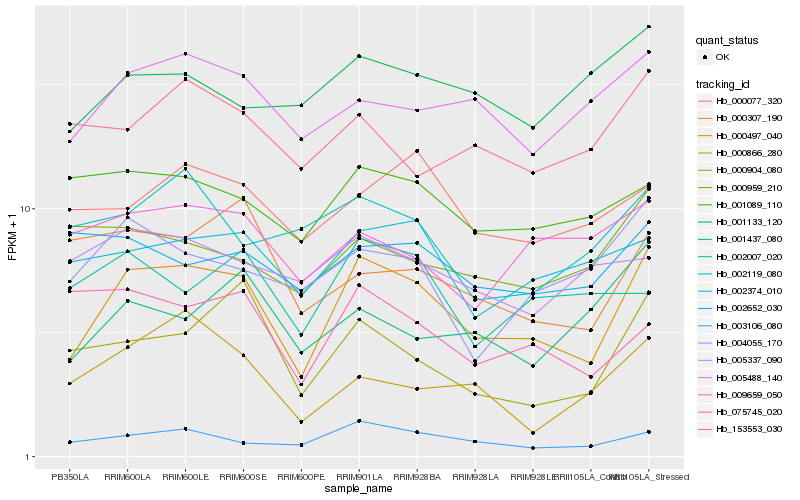
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000497_040 |
0.0 |
- |
- |
BnaC09g09960D [Brassica napus] |
| 2 |
Hb_003106_080 |
0.1332584861 |
- |
- |
cell division cycle, putative [Ricinus communis] |
| 3 |
Hb_005488_140 |
0.1530620996 |
- |
- |
30S ribosomal protein S5, putative [Ricinus communis] |
| 4 |
Hb_000904_080 |
0.1555951724 |
- |
- |
alpha-amylase-like protein [Vitis labrusca x Vitis vinifera] |
| 5 |
Hb_002374_010 |
0.1582505147 |
- |
- |
PREDICTED: protein translocase subunit SECA2, chloroplastic [Jatropha curcas] |
| 6 |
Hb_004055_170 |
0.1634719253 |
- |
- |
nucleosome assembly protein, putative [Ricinus communis] |
| 7 |
Hb_000077_320 |
0.1649258223 |
- |
- |
PREDICTED: lanC-like protein GCL2 [Jatropha curcas] |
| 8 |
Hb_001089_110 |
0.1678261211 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001437_080 |
0.1703477529 |
transcription factor |
TF Family: TCP |
hypothetical protein JCGZ_05472 [Jatropha curcas] |
| 10 |
Hb_001133_120 |
0.1707037157 |
- |
- |
PREDICTED: uncharacterized protein LOC105629988 [Jatropha curcas] |
| 11 |
Hb_153553_030 |
0.1715762824 |
- |
- |
PREDICTED: zinc finger protein GIS2 [Jatropha curcas] |
| 12 |
Hb_075745_020 |
0.1717087942 |
- |
- |
PREDICTED: uncharacterized protein LOC105649606 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002119_080 |
0.1730423586 |
- |
- |
PREDICTED: 54S ribosomal protein L24, mitochondrial [Jatropha curcas] |
| 14 |
Hb_009659_050 |
0.1740750067 |
- |
- |
PREDICTED: putative hydrolase C777.06c isoform X2 [Jatropha curcas] |
| 15 |
Hb_002007_020 |
0.1750952066 |
- |
- |
PREDICTED: ATP-dependent DNA helicase Q-like SIM [Jatropha curcas] |
| 16 |
Hb_000866_280 |
0.1756983835 |
- |
- |
PREDICTED: seven transmembrane domain-containing tyrosine-protein kinase 1-like [Jatropha curcas] |
| 17 |
Hb_000307_190 |
0.1761856958 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g20300, mitochondrial-like [Populus euphratica] |
| 18 |
Hb_000959_210 |
0.1762281031 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 27 [Jatropha curcas] |
| 19 |
Hb_002652_030 |
0.1771045591 |
- |
- |
PREDICTED: U3 small nucleolar RNA-interacting protein 2-like [Jatropha curcas] |
| 20 |
Hb_005337_090 |
0.1774182678 |
- |
- |
PREDICTED: serine/threonine-protein kinase TOUSLED-like [Jatropha curcas] |