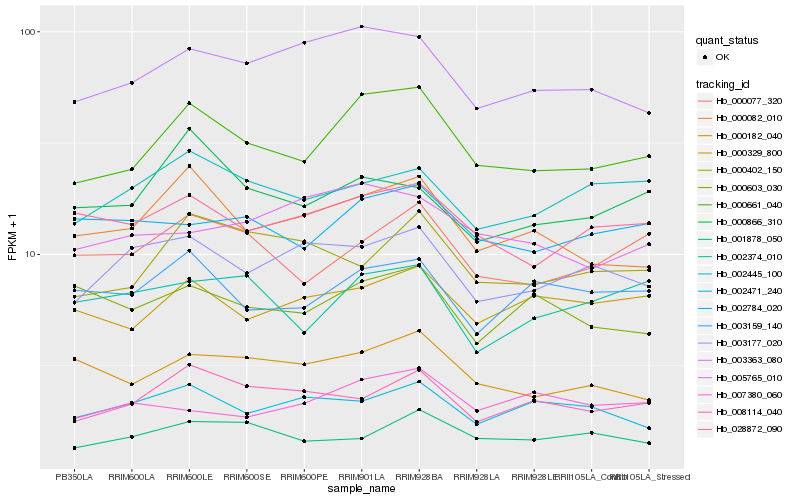
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000661_040 |
0.0 |
- |
- |
PREDICTED: dihydroxy-acid dehydratase, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000082_010 |
0.0883849088 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RGLG2 [Jatropha curcas] |
| 3 |
Hb_003363_080 |
0.0919578511 |
- |
- |
eukaryotic translation elongation factor 1B gamma-subunit [Hevea brasiliensis] |
| 4 |
Hb_028872_090 |
0.097056607 |
- |
- |
PREDICTED: urease [Jatropha curcas] |
| 5 |
Hb_000077_320 |
0.0970595247 |
- |
- |
PREDICTED: lanC-like protein GCL2 [Jatropha curcas] |
| 6 |
Hb_000329_800 |
0.1017285224 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_008114_040 |
0.1043578348 |
- |
- |
Fanconi anemia group D2 [Gossypium arboreum] |
| 8 |
Hb_000182_040 |
0.1046641469 |
transcription factor |
TF Family: SNF2 |
PREDICTED: protein CHROMATIN REMODELING 24 [Jatropha curcas] |
| 9 |
Hb_002784_020 |
0.1056164974 |
- |
- |
PREDICTED: probable protein arginine N-methyltransferase 3 [Jatropha curcas] |
| 10 |
Hb_003159_140 |
0.1063325363 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_003177_020 |
0.1081275257 |
- |
- |
diphthine synthase, putative [Ricinus communis] |
| 12 |
Hb_002445_100 |
0.1091364786 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 5 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000402_150 |
0.1096878183 |
- |
- |
PREDICTED: uncharacterized protein LOC105644650 isoform X2 [Jatropha curcas] |
| 14 |
Hb_001878_050 |
0.1123239982 |
- |
- |
PREDICTED: uncharacterized protein LOC105646110 isoform X3 [Jatropha curcas] |
| 15 |
Hb_005765_010 |
0.1129008255 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At1g63170 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000866_310 |
0.1163087714 |
- |
- |
PREDICTED: neural Wiskott-Aldrich syndrome protein [Jatropha curcas] |
| 17 |
Hb_000603_030 |
0.1166092493 |
- |
- |
Ribonuclease III, putative [Ricinus communis] |
| 18 |
Hb_007380_060 |
0.1170144314 |
- |
- |
hypothetical protein POPTR_0006s00280g [Populus trichocarpa] |
| 19 |
Hb_002374_010 |
0.1173054859 |
- |
- |
PREDICTED: protein translocase subunit SECA2, chloroplastic [Jatropha curcas] |
| 20 |
Hb_002471_240 |
0.1176707087 |
- |
- |
PREDICTED: protein POLLEN DEFECTIVE IN GUIDANCE 1 isoform X1 [Jatropha curcas] |