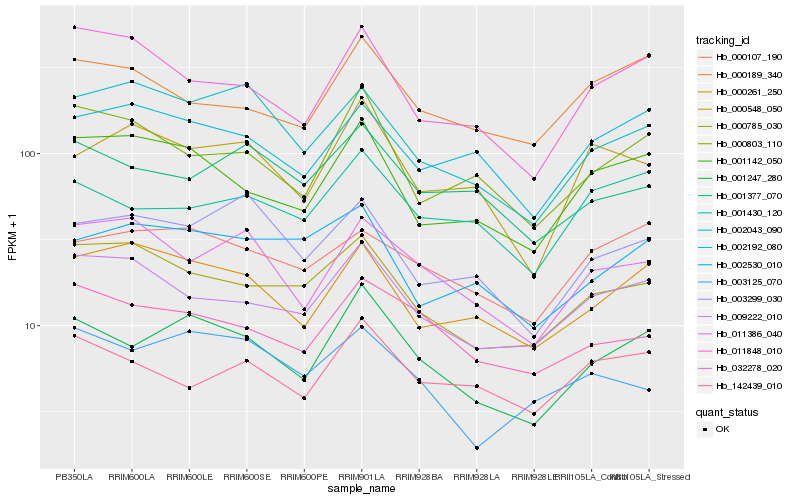
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001247_280 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632071 [Jatropha curcas] |
| 2 |
Hb_001142_050 |
0.1170266364 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 12 [Jatropha curcas] |
| 3 |
Hb_032278_020 |
0.1211261282 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 16 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000261_250 |
0.1243333103 |
- |
- |
elongation factor 1-alpha [Lepidium sativum] |
| 5 |
Hb_001430_120 |
0.1272937381 |
- |
- |
PREDICTED: DNA polymerase delta subunit 4 [Jatropha curcas] |
| 6 |
Hb_000803_110 |
0.1273561833 |
- |
- |
PREDICTED: RNA-binding protein 8A [Jatropha curcas] |
| 7 |
Hb_009222_010 |
0.1284911012 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 8 |
Hb_000785_030 |
0.1313563287 |
- |
- |
hypothetical protein RCOM_0808030 [Ricinus communis] |
| 9 |
Hb_011848_010 |
0.1335770284 |
- |
- |
PREDICTED: 40S ribosomal protein S4-3 [Jatropha curcas] |
| 10 |
Hb_000107_190 |
0.134505858 |
- |
- |
50S ribosomal protein L20 [Hevea brasiliensis] |
| 11 |
Hb_000548_050 |
0.1355068717 |
- |
- |
hypothetical protein POPTR_0008s12570g [Populus trichocarpa] |
| 12 |
Hb_001377_070 |
0.1357281561 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002043_090 |
0.1375465854 |
- |
- |
hypothetical protein JCGZ_24811 [Jatropha curcas] |
| 14 |
Hb_142439_010 |
0.1376094548 |
- |
- |
PREDICTED: F-box/kelch-repeat protein OR23 [Jatropha curcas] |
| 15 |
Hb_000189_340 |
0.1377038555 |
- |
- |
PREDICTED: 40S ribosomal protein S3a-1 [Jatropha curcas] |
| 16 |
Hb_003299_030 |
0.137840658 |
- |
- |
PREDICTED: uncharacterized protein At1g27050 isoform X1 [Jatropha curcas] |
| 17 |
Hb_002192_080 |
0.138150732 |
- |
- |
hypothetical protein B456_013G069200 [Gossypium raimondii] |
| 18 |
Hb_003125_070 |
0.1382678336 |
- |
- |
PREDICTED: serine/threonine-protein kinase rio1-like [Jatropha curcas] |
| 19 |
Hb_011386_040 |
0.1390632486 |
- |
- |
PREDICTED: uncharacterized protein LOC105639243 isoform X2 [Jatropha curcas] |
| 20 |
Hb_002530_010 |
0.1399547801 |
- |
- |
hypothetical protein EUGRSUZ_C02580 [Eucalyptus grandis] |