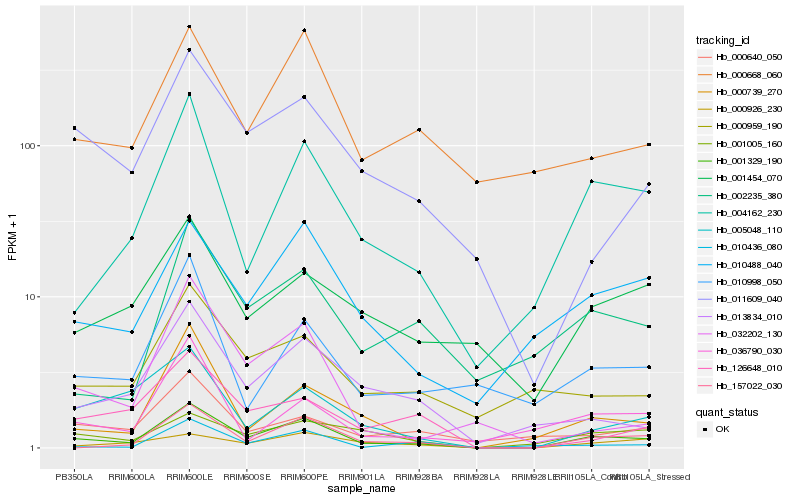
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001329_190 |
0.0 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 2 |
Hb_004162_230 |
0.1908690124 |
- |
- |
PREDICTED: histone H2B [Jatropha curcas] |
| 3 |
Hb_157022_030 |
0.1980832936 |
transcription factor |
TF Family: NF-YB |
ccaat-binding transcription factor subunit A, putative [Ricinus communis] |
| 4 |
Hb_000739_270 |
0.2046636793 |
- |
- |
PREDICTED: serine/threonine-protein kinase Aurora-3 [Jatropha curcas] |
| 5 |
Hb_010488_040 |
0.2060957407 |
- |
- |
PREDICTED: phosphomannomutase [Populus euphratica] |
| 6 |
Hb_011609_040 |
0.2083359905 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 40 [Jatropha curcas] |
| 7 |
Hb_013834_010 |
0.2158950432 |
- |
- |
PREDICTED: kinesin-like protein NACK2 [Jatropha curcas] |
| 8 |
Hb_005048_110 |
0.2178526691 |
- |
- |
- |
| 9 |
Hb_000668_060 |
0.2221660462 |
- |
- |
Pollen-specific protein C13 precursor, putative [Ricinus communis] |
| 10 |
Hb_126648_010 |
0.2268424855 |
- |
- |
PREDICTED: vacuolar protein-sorting-associated protein 33 homolog [Jatropha curcas] |
| 11 |
Hb_001005_160 |
0.2277754832 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_010998_050 |
0.2300566668 |
- |
- |
PREDICTED: kinesin-like protein KIF22 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000926_230 |
0.2304021299 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000959_190 |
0.2305671999 |
- |
- |
PREDICTED: crossover junction endonuclease EME1B-like [Jatropha curcas] |
| 15 |
Hb_032202_130 |
0.2357508237 |
- |
- |
PREDICTED: uncharacterized protein LOC105643062 [Jatropha curcas] |
| 16 |
Hb_000640_050 |
0.2370844752 |
- |
- |
DNA replication licensing factor MCM7, putative [Ricinus communis] |
| 17 |
Hb_002235_380 |
0.2375234143 |
- |
- |
hypothetical protein JCGZ_19860 [Jatropha curcas] |
| 18 |
Hb_001454_070 |
0.238563263 |
- |
- |
Uveal autoantigen with coiled-coil domains and ankyrin repeats isoform 1 [Theobroma cacao] |
| 19 |
Hb_010436_080 |
0.240106803 |
- |
- |
PREDICTED: DNA replication licensing factor MCM5 [Jatropha curcas] |
| 20 |
Hb_036790_030 |
0.240373718 |
- |
- |
PREDICTED: UPF0544 protein C5orf45 homolog [Jatropha curcas] |