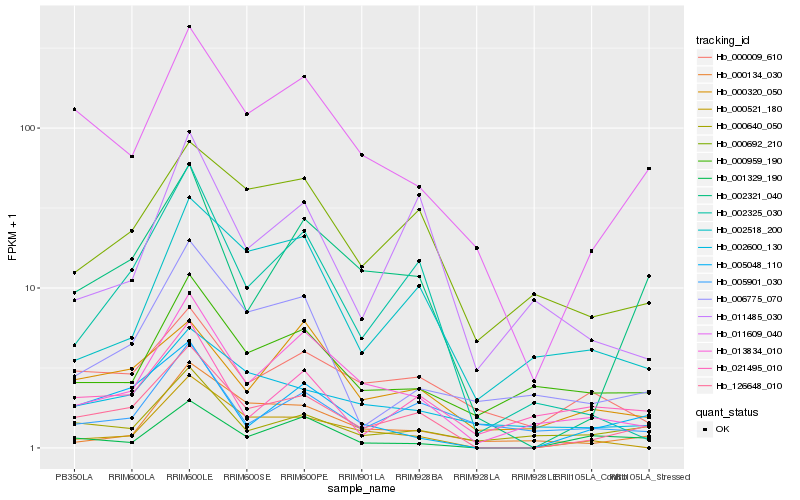
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_126648_010 |
0.0 |
- |
- |
PREDICTED: vacuolar protein-sorting-associated protein 33 homolog [Jatropha curcas] |
| 2 |
Hb_002321_040 |
0.1464709452 |
- |
- |
hypothetical protein POPTR_0019s14030g [Populus trichocarpa] |
| 3 |
Hb_011609_040 |
0.1641397471 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 40 [Jatropha curcas] |
| 4 |
Hb_005048_110 |
0.1890123356 |
- |
- |
- |
| 5 |
Hb_013834_010 |
0.1910464861 |
- |
- |
PREDICTED: kinesin-like protein NACK2 [Jatropha curcas] |
| 6 |
Hb_002325_030 |
0.1971609611 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 2 [Jatropha curcas] |
| 7 |
Hb_000692_210 |
0.2046632236 |
- |
- |
PREDICTED: 24-methylenesterol C-methyltransferase 2 [Jatropha curcas] |
| 8 |
Hb_000640_050 |
0.2104183282 |
- |
- |
DNA replication licensing factor MCM7, putative [Ricinus communis] |
| 9 |
Hb_002600_130 |
0.2141986741 |
- |
- |
PREDICTED: uncharacterized protein LOC105646808 [Jatropha curcas] |
| 10 |
Hb_000521_180 |
0.2178248774 |
- |
- |
PREDICTED: uncharacterized protein LOC105650335 [Jatropha curcas] |
| 11 |
Hb_000959_190 |
0.222536166 |
- |
- |
PREDICTED: crossover junction endonuclease EME1B-like [Jatropha curcas] |
| 12 |
Hb_000320_050 |
0.2249951914 |
- |
- |
bcr-associated protein, bap, putative [Ricinus communis] |
| 13 |
Hb_006775_070 |
0.2258675748 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 14 |
Hb_000009_610 |
0.2266272511 |
- |
- |
PREDICTED: uncharacterized protein LOC105639638 isoform X2 [Jatropha curcas] |
| 15 |
Hb_001329_190 |
0.2268424855 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 16 |
Hb_002518_200 |
0.2315177085 |
- |
- |
Proliferating cell nuclear antigen [Theobroma cacao] |
| 17 |
Hb_021495_010 |
0.231935559 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_011485_030 |
0.2325255719 |
- |
- |
PREDICTED: uncharacterized protein LOC105647814 [Jatropha curcas] |
| 19 |
Hb_000134_030 |
0.2339388135 |
- |
- |
PREDICTED: sister chromatid cohesion 1 protein 3-like [Jatropha curcas] |
| 20 |
Hb_005901_030 |
0.2350928683 |
- |
- |
PREDICTED: phragmoplast orienting kinesin 2 isoform X1 [Jatropha curcas] |