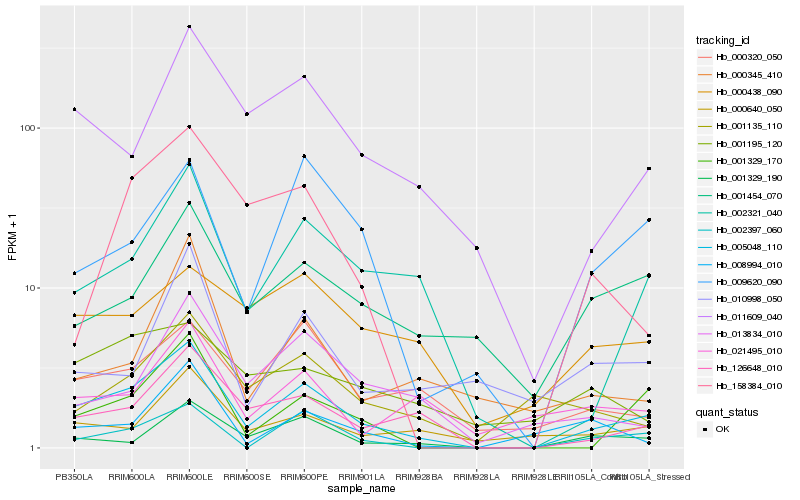
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005048_110 |
0.0 |
- |
- |
- |
| 2 |
Hb_126648_010 |
0.1890123356 |
- |
- |
PREDICTED: vacuolar protein-sorting-associated protein 33 homolog [Jatropha curcas] |
| 3 |
Hb_002321_040 |
0.191368498 |
- |
- |
hypothetical protein POPTR_0019s14030g [Populus trichocarpa] |
| 4 |
Hb_011609_040 |
0.2108159497 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 40 [Jatropha curcas] |
| 5 |
Hb_001329_170 |
0.2122716157 |
- |
- |
Chlorophyll a-b binding protein, chloroplastic [Aegilops tauschii] |
| 6 |
Hb_001329_190 |
0.2178526691 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 7 |
Hb_002397_060 |
0.2197061517 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_009620_090 |
0.2304286779 |
- |
- |
hypothetical protein L484_000980 [Morus notabilis] |
| 9 |
Hb_000640_050 |
0.233890371 |
- |
- |
DNA replication licensing factor MCM7, putative [Ricinus communis] |
| 10 |
Hb_000320_050 |
0.2361800229 |
- |
- |
bcr-associated protein, bap, putative [Ricinus communis] |
| 11 |
Hb_013834_010 |
0.2432319401 |
- |
- |
PREDICTED: kinesin-like protein NACK2 [Jatropha curcas] |
| 12 |
Hb_001454_070 |
0.2440895992 |
- |
- |
Uveal autoantigen with coiled-coil domains and ankyrin repeats isoform 1 [Theobroma cacao] |
| 13 |
Hb_001135_110 |
0.2506053055 |
- |
- |
- |
| 14 |
Hb_158384_010 |
0.2512456686 |
- |
- |
poly(A) polymerase, putative [Ricinus communis] |
| 15 |
Hb_021495_010 |
0.2512768472 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000438_090 |
0.2527469315 |
- |
- |
PREDICTED: mini-chromosome maintenance complex-binding protein [Jatropha curcas] |
| 17 |
Hb_000345_410 |
0.2549196302 |
- |
- |
PREDICTED: kinesin-like protein KIFC3 [Jatropha curcas] |
| 18 |
Hb_010998_050 |
0.2594873521 |
- |
- |
PREDICTED: kinesin-like protein KIF22 isoform X2 [Jatropha curcas] |
| 19 |
Hb_008994_010 |
0.2603117394 |
- |
- |
Pectin lyase-like superfamily protein isoform 1 [Theobroma cacao] |
| 20 |
Hb_001195_120 |
0.261794038 |
- |
- |
oxidoreductase, putative [Ricinus communis] |