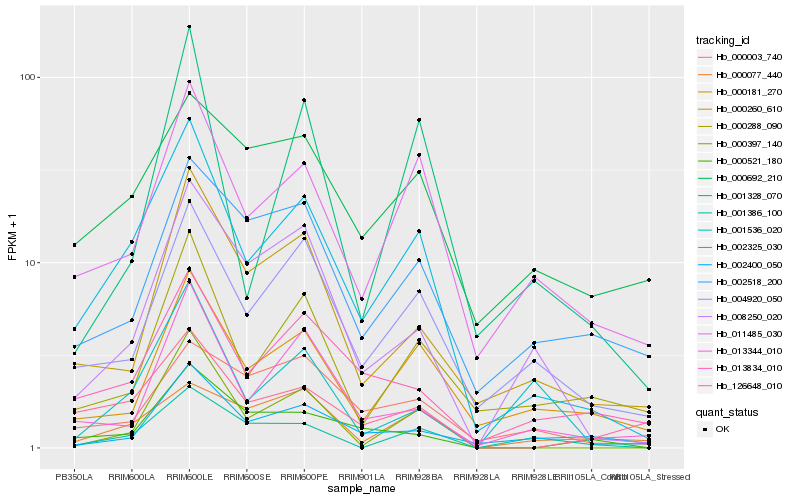
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002325_030 |
0.0 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 2 [Jatropha curcas] |
| 2 |
Hb_011485_030 |
0.1669675211 |
- |
- |
PREDICTED: uncharacterized protein LOC105647814 [Jatropha curcas] |
| 3 |
Hb_004920_050 |
0.1765941213 |
- |
- |
DNA replication licensing factor MCM7, putative [Ricinus communis] |
| 4 |
Hb_000397_140 |
0.1903889642 |
transcription factor |
TF Family: PLATZ |
hypothetical protein JCGZ_16980 [Jatropha curcas] |
| 5 |
Hb_013834_010 |
0.1913136118 |
- |
- |
PREDICTED: kinesin-like protein NACK2 [Jatropha curcas] |
| 6 |
Hb_126648_010 |
0.1971609611 |
- |
- |
PREDICTED: vacuolar protein-sorting-associated protein 33 homolog [Jatropha curcas] |
| 7 |
Hb_008250_020 |
0.197506691 |
- |
- |
PREDICTED: U-box domain-containing protein 15-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_000260_610 |
0.1986346177 |
- |
- |
PREDICTED: cell division cycle 20.1, cofactor of APC complex-like [Jatropha curcas] |
| 9 |
Hb_000181_270 |
0.1989827518 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 10 |
Hb_013344_010 |
0.2016975834 |
- |
- |
PREDICTED: microtubule-associated protein TORTIFOLIA1 [Jatropha curcas] |
| 11 |
Hb_000288_090 |
0.2060216007 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 12 |
Hb_000521_180 |
0.2089676453 |
- |
- |
PREDICTED: uncharacterized protein LOC105650335 [Jatropha curcas] |
| 13 |
Hb_001386_100 |
0.2174576093 |
transcription factor |
TF Family: M-type |
PREDICTED: MADS-box transcription factor PHERES 1-like [Jatropha curcas] |
| 14 |
Hb_002400_050 |
0.2223936775 |
- |
- |
PREDICTED: uncharacterized protein LOC105637632 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000003_740 |
0.2224358405 |
- |
- |
PREDICTED: uncharacterized protein LOC105631539 [Jatropha curcas] |
| 16 |
Hb_001328_070 |
0.2227259836 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 13 [Jatropha curcas] |
| 17 |
Hb_000692_210 |
0.2235578845 |
- |
- |
PREDICTED: 24-methylenesterol C-methyltransferase 2 [Jatropha curcas] |
| 18 |
Hb_002518_200 |
0.2245544019 |
- |
- |
Proliferating cell nuclear antigen [Theobroma cacao] |
| 19 |
Hb_000077_440 |
0.229563645 |
- |
- |
PREDICTED: ELMO domain-containing protein A isoform X1 [Jatropha curcas] |
| 20 |
Hb_001536_020 |
0.2309249807 |
transcription factor |
TF Family: GRF |
PREDICTED: uncharacterized protein LOC105631006 [Jatropha curcas] |