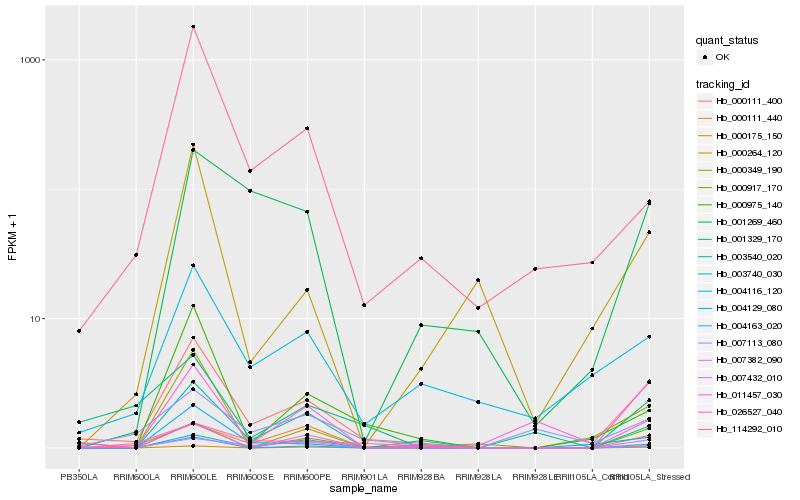
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000111_400 |
0.0 |
- |
- |
hypothetical protein POPTR_0003s19310g [Populus trichocarpa] |
| 2 |
Hb_004163_020 |
0.2278559969 |
- |
- |
- |
| 3 |
Hb_000349_190 |
0.2595355475 |
transcription factor |
TF Family: GRF |
hypothetical protein POPTR_0001s08310g [Populus trichocarpa] |
| 4 |
Hb_004116_120 |
0.2621365582 |
- |
- |
PREDICTED: protein EARLY FLOWERING 4 [Jatropha curcas] |
| 5 |
Hb_004129_080 |
0.2696009119 |
- |
- |
PREDICTED: putative DNA-binding protein ESCAROLA [Prunus mume] |
| 6 |
Hb_026527_040 |
0.2703747157 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001329_170 |
0.2743430337 |
- |
- |
Chlorophyll a-b binding protein, chloroplastic [Aegilops tauschii] |
| 8 |
Hb_000111_440 |
0.2788782278 |
- |
- |
- |
| 9 |
Hb_000975_140 |
0.2790703273 |
- |
- |
lactoylglutathione lyase family protein [Populus trichocarpa] |
| 10 |
Hb_114292_010 |
0.2810666419 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_003740_030 |
0.2824209716 |
transcription factor |
TF Family: RAV |
DNA-binding protein RAV1, putative [Ricinus communis] |
| 12 |
Hb_001269_460 |
0.2827448338 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 40 [Jatropha curcas] |
| 13 |
Hb_000175_150 |
0.2855000065 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein B456_009G311700 [Gossypium raimondii] |
| 14 |
Hb_000917_170 |
0.2862070313 |
- |
- |
PREDICTED: clavaminate synthase-like protein At3g21360 [Jatropha curcas] |
| 15 |
Hb_007382_090 |
0.2875428527 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 16 |
Hb_003540_020 |
0.2898279582 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_007432_010 |
0.291153382 |
- |
- |
hypothetical protein B456_004G088300 [Gossypium raimondii] |
| 18 |
Hb_011457_030 |
0.2936706348 |
- |
- |
type I inositol polyphosphate 5-phosphatase, putative [Ricinus communis] |
| 19 |
Hb_000264_120 |
0.2968142103 |
- |
- |
Stem-specific protein TSJT1, putative [Ricinus communis] |
| 20 |
Hb_007113_080 |
0.2994675331 |
- |
- |
PREDICTED: uncharacterized protein LOC105639553 isoform X1 [Jatropha curcas] |