| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
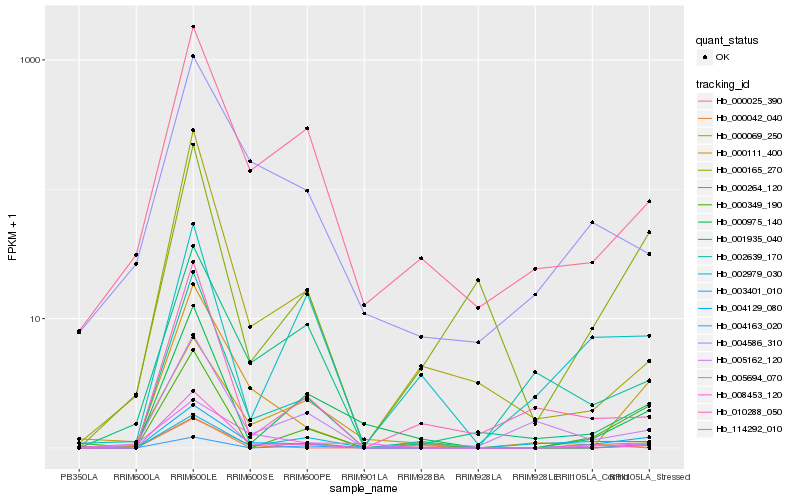
Hb_004129_080 |
0.0 |
- |
- |
PREDICTED: putative DNA-binding protein ESCAROLA [Prunus mume] |
| 2 |
Hb_000349_190 |
0.2125296422 |
transcription factor |
TF Family: GRF |
hypothetical protein POPTR_0001s08310g [Populus trichocarpa] |
| 3 |
Hb_000264_120 |
0.2317110521 |
- |
- |
Stem-specific protein TSJT1, putative [Ricinus communis] |
| 4 |
Hb_002639_170 |
0.2476065717 |
- |
- |
carboxy-lyase, putative [Ricinus communis] |
| 5 |
Hb_004586_310 |
0.2488527702 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_114292_010 |
0.2546648519 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000975_140 |
0.261217014 |
- |
- |
lactoylglutathione lyase family protein [Populus trichocarpa] |
| 8 |
Hb_003401_010 |
0.2632163432 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 9 |
Hb_004163_020 |
0.2649041049 |
- |
- |
- |
| 10 |
Hb_000111_400 |
0.2696009119 |
- |
- |
hypothetical protein POPTR_0003s19310g [Populus trichocarpa] |
| 11 |
Hb_005162_120 |
0.2706924355 |
- |
- |
ferric reductase oxidase [Manihot esculenta] |
| 12 |
Hb_001935_040 |
0.2756808737 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 8 [Jatropha curcas] |
| 13 |
Hb_008453_120 |
0.2772585583 |
transcription factor |
TF Family: bZIP |
hypothetical protein POPTR_0005s26480g [Populus trichocarpa] |
| 14 |
Hb_005694_070 |
0.2782922253 |
- |
- |
kinase, putative [Ricinus communis] |
| 15 |
Hb_000042_040 |
0.2812651145 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g23880-like [Jatropha curcas] |
| 16 |
Hb_002979_030 |
0.2839530816 |
- |
- |
PREDICTED: serine/threonine-protein kinase Aurora-1 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000025_390 |
0.2860824253 |
- |
- |
PREDICTED: putative DNA-binding protein ESCAROLA [Populus euphratica] |
| 18 |
Hb_000165_270 |
0.2864062741 |
transcription factor |
TF Family: Tify |
JAZ10 [Hevea brasiliensis] |
| 19 |
Hb_010288_050 |
0.288892741 |
- |
- |
PREDICTED: uncharacterized protein LOC105642604 [Jatropha curcas] |
| 20 |
Hb_000069_250 |
0.2914191988 |
- |
- |
hypothetical protein JCGZ_14250 [Jatropha curcas] |