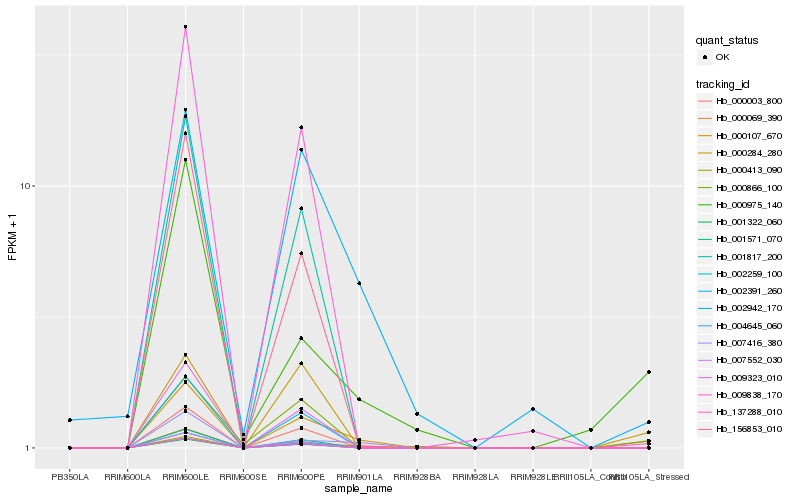
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000107_670 |
0.0 |
transcription factor |
TF Family: MIKC |
floral homeotic protein AGAMOUS [Jatropha curcas] |
| 2 |
Hb_000069_390 |
0.1963480053 |
- |
- |
PREDICTED: probable F-box protein At3g61730 [Jatropha curcas] |
| 3 |
Hb_000975_140 |
0.2071128487 |
- |
- |
lactoylglutathione lyase family protein [Populus trichocarpa] |
| 4 |
Hb_007416_380 |
0.2092779036 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 3 [Jatropha curcas] |
| 5 |
Hb_002942_170 |
0.215477869 |
- |
- |
acyl carrier protein, putative [Ricinus communis] |
| 6 |
Hb_000866_100 |
0.2277443273 |
- |
- |
PREDICTED: protein PAIR1 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000284_280 |
0.2310220232 |
- |
- |
- |
| 8 |
Hb_156853_010 |
0.236298607 |
- |
- |
PREDICTED: vinorine synthase-like [Populus euphratica] |
| 9 |
Hb_009838_170 |
0.2364314332 |
- |
- |
Arylacetamide deacetylase, putative [Ricinus communis] |
| 10 |
Hb_002259_100 |
0.2373153462 |
- |
- |
PREDICTED: uncharacterized protein LOC105630152 isoform X1 [Jatropha curcas] |
| 11 |
Hb_004645_060 |
0.2373543687 |
- |
- |
PREDICTED: uncharacterized protein LOC105630948 [Jatropha curcas] |
| 12 |
Hb_001817_200 |
0.2374257901 |
- |
- |
hypothetical protein CISIN_1g037825mg [Citrus sinensis] |
| 13 |
Hb_002391_260 |
0.2374885478 |
- |
- |
hypothetical protein RCOM_0017280 [Ricinus communis] |
| 14 |
Hb_001571_070 |
0.2375209721 |
- |
- |
PREDICTED: probable inactive receptor kinase At2g26730 [Jatropha curcas] |
| 15 |
Hb_000413_090 |
0.2376695792 |
- |
- |
PREDICTED: transcription factor DYT1 [Jatropha curcas] |
| 16 |
Hb_009323_010 |
0.2378148107 |
- |
- |
hypothetical protein POPTR_0008s03350g, partial [Populus trichocarpa] |
| 17 |
Hb_007552_030 |
0.238047137 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_21064 [Jatropha curcas] |
| 18 |
Hb_001322_060 |
0.2380880608 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 19 |
Hb_000003_800 |
0.2380894339 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_137288_010 |
0.2381656712 |
- |
- |
hypothetical protein B456_003G055000 [Gossypium raimondii] |