| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
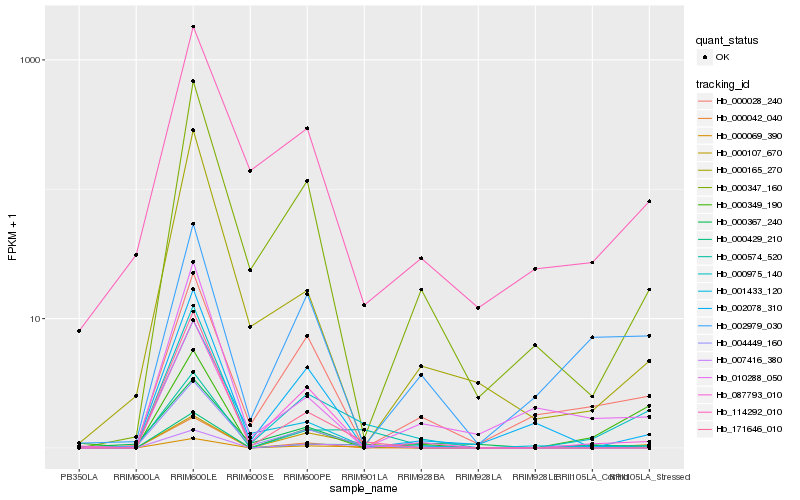
Hb_000975_140 |
0.0 |
- |
- |
lactoylglutathione lyase family protein [Populus trichocarpa] |
| 2 |
Hb_000042_040 |
0.1835599585 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g23880-like [Jatropha curcas] |
| 3 |
Hb_000347_160 |
0.201007776 |
transcription factor |
TF Family: Tify |
JAZ2 [Hevea brasiliensis] |
| 4 |
Hb_087793_010 |
0.201490796 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000107_670 |
0.2071128487 |
transcription factor |
TF Family: MIKC |
floral homeotic protein AGAMOUS [Jatropha curcas] |
| 6 |
Hb_114292_010 |
0.2145024988 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_004449_160 |
0.2212918815 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 8 |
Hb_000028_240 |
0.2276200529 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000349_190 |
0.2315659122 |
transcription factor |
TF Family: GRF |
hypothetical protein POPTR_0001s08310g [Populus trichocarpa] |
| 10 |
Hb_171646_010 |
0.2330360473 |
- |
- |
hypothetical protein B456_012G144300, partial [Gossypium raimondii] |
| 11 |
Hb_000069_390 |
0.2339170148 |
- |
- |
PREDICTED: probable F-box protein At3g61730 [Jatropha curcas] |
| 12 |
Hb_000367_240 |
0.2347037534 |
- |
- |
hypothetical protein JCGZ_11682 [Jatropha curcas] |
| 13 |
Hb_002078_310 |
0.2362943174 |
- |
- |
PREDICTED: uncharacterized protein LOC105645005 [Jatropha curcas] |
| 14 |
Hb_000574_520 |
0.2373785991 |
- |
- |
PREDICTED: uncharacterized protein LOC105634842 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000165_270 |
0.2375599158 |
transcription factor |
TF Family: Tify |
JAZ10 [Hevea brasiliensis] |
| 16 |
Hb_001433_120 |
0.2384016924 |
- |
- |
hypothetical protein CICLE_v10003574mg, partial [Citrus clementina] |
| 17 |
Hb_010288_050 |
0.2384688358 |
- |
- |
PREDICTED: uncharacterized protein LOC105642604 [Jatropha curcas] |
| 18 |
Hb_007416_380 |
0.24007704 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 3 [Jatropha curcas] |
| 19 |
Hb_000429_210 |
0.2401635204 |
transcription factor |
TF Family: ERF |
hypothetical protein RCOM_0089590 [Ricinus communis] |
| 20 |
Hb_002979_030 |
0.2420828293 |
- |
- |
PREDICTED: serine/threonine-protein kinase Aurora-1 isoform X2 [Jatropha curcas] |