| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
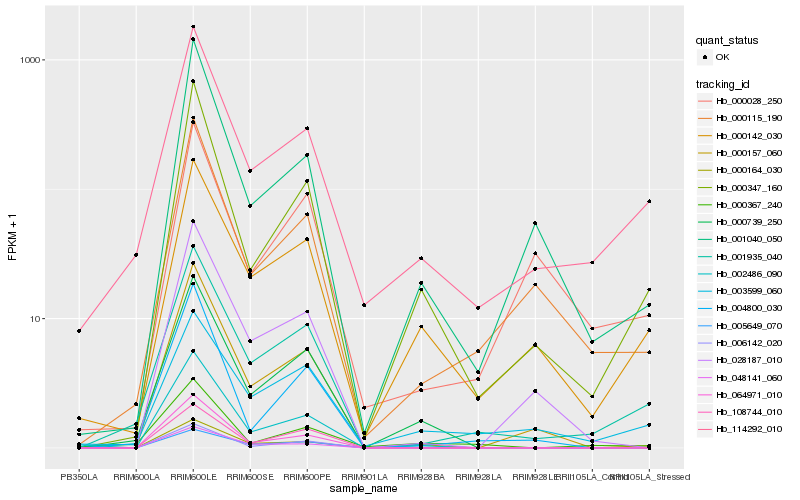
Hb_001935_040 |
0.0 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 8 [Jatropha curcas] |
| 2 |
Hb_114292_010 |
0.1102376854 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000115_190 |
0.1447008652 |
- |
- |
PREDICTED: uncharacterized protein LOC105642039 [Jatropha curcas] |
| 4 |
Hb_000142_030 |
0.1511603007 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 5 |
Hb_000347_160 |
0.1532489673 |
transcription factor |
TF Family: Tify |
JAZ2 [Hevea brasiliensis] |
| 6 |
Hb_000157_060 |
0.1551771292 |
- |
- |
PREDICTED: polyamine oxidase 1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_003599_060 |
0.1551871137 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 8 |
Hb_028187_010 |
0.1689864114 |
- |
- |
hypothetical protein JCGZ_23781 [Jatropha curcas] |
| 9 |
Hb_000164_030 |
0.1703498726 |
- |
- |
PREDICTED: RING-H2 finger protein ATL14-like [Zea mays] |
| 10 |
Hb_000739_250 |
0.172324951 |
- |
- |
PREDICTED: uncharacterized protein LOC105120821 [Populus euphratica] |
| 11 |
Hb_064971_010 |
0.1753546139 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 12 |
Hb_001040_050 |
0.1760660394 |
- |
- |
Stem-specific protein TSJT1, putative [Ricinus communis] |
| 13 |
Hb_006142_020 |
0.1768073811 |
- |
- |
JHL25P11.12 [Jatropha curcas] |
| 14 |
Hb_005649_070 |
0.177754592 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_048141_060 |
0.1793169929 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein ZAT3 [Populus euphratica] |
| 16 |
Hb_004800_030 |
0.1802169439 |
transcription factor |
TF Family: OFP |
PREDICTED: transcription repressor OFP7-like [Jatropha curcas] |
| 17 |
Hb_108744_010 |
0.1816695173 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 18 |
Hb_000367_240 |
0.1819793296 |
- |
- |
hypothetical protein JCGZ_11682 [Jatropha curcas] |
| 19 |
Hb_000028_250 |
0.1845624879 |
- |
- |
PREDICTED: nudix hydrolase 4 [Jatropha curcas] |
| 20 |
Hb_002486_090 |
0.1854772966 |
- |
- |
hypothetical protein POPTR_0005s06910g [Populus trichocarpa] |