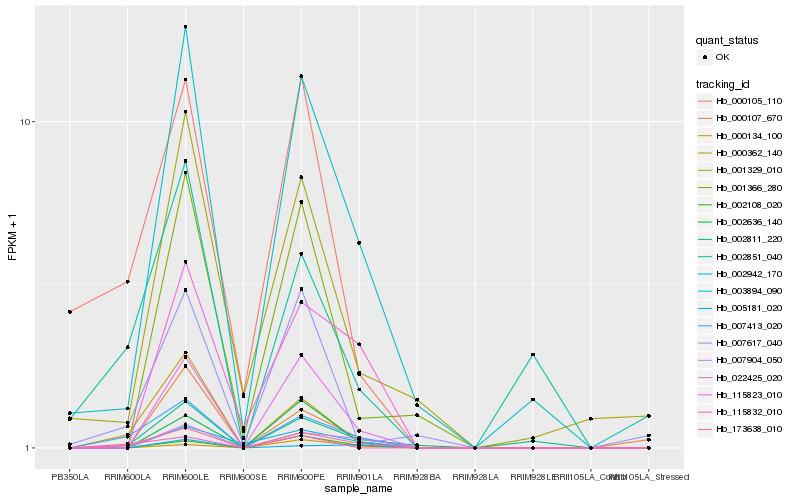
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_173638_010 |
0.0 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 2 |
Hb_007413_020 |
0.077237368 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000362_140 |
0.1707321867 |
transcription factor |
TF Family: bZIP |
PREDICTED: basic leucine zipper 61-like [Jatropha curcas] |
| 4 |
Hb_002942_170 |
0.185664066 |
- |
- |
acyl carrier protein, putative [Ricinus communis] |
| 5 |
Hb_022425_020 |
0.1921373276 |
- |
- |
- |
| 6 |
Hb_007904_050 |
0.1989239607 |
- |
- |
hypothetical protein JCGZ_17883 [Jatropha curcas] |
| 7 |
Hb_002811_220 |
0.2288793041 |
transcription factor |
TF Family: Trihelix |
transcription factor, putative [Ricinus communis] |
| 8 |
Hb_115823_010 |
0.2295769133 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_002636_140 |
0.2347565747 |
- |
- |
PREDICTED: GDSL esterase/lipase At1g71691-like [Populus euphratica] |
| 10 |
Hb_002108_020 |
0.2368841286 |
- |
- |
hypothetical protein JCGZ_16012 [Jatropha curcas] |
| 11 |
Hb_000105_110 |
0.2408622844 |
- |
- |
hypothetical protein, partial [Fervidibacteria bacterium SCGC AAA471-D06] |
| 12 |
Hb_001366_280 |
0.2582609962 |
- |
- |
PREDICTED: uncharacterized protein LOC105642732 [Jatropha curcas] |
| 13 |
Hb_007617_040 |
0.2596318954 |
- |
- |
PREDICTED: syntaxin-112-like [Jatropha curcas] |
| 14 |
Hb_115832_010 |
0.260963391 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 15 |
Hb_002851_040 |
0.2615294316 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 5 [Jatropha curcas] |
| 16 |
Hb_003894_090 |
0.2621536198 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001329_010 |
0.2651671696 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 18 |
Hb_000107_670 |
0.2688264844 |
transcription factor |
TF Family: MIKC |
floral homeotic protein AGAMOUS [Jatropha curcas] |
| 19 |
Hb_000134_100 |
0.2832869868 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Prunus mume] |
| 20 |
Hb_005181_020 |
0.2849980574 |
- |
- |
Pectinesterase-2 precursor, putative [Ricinus communis] |