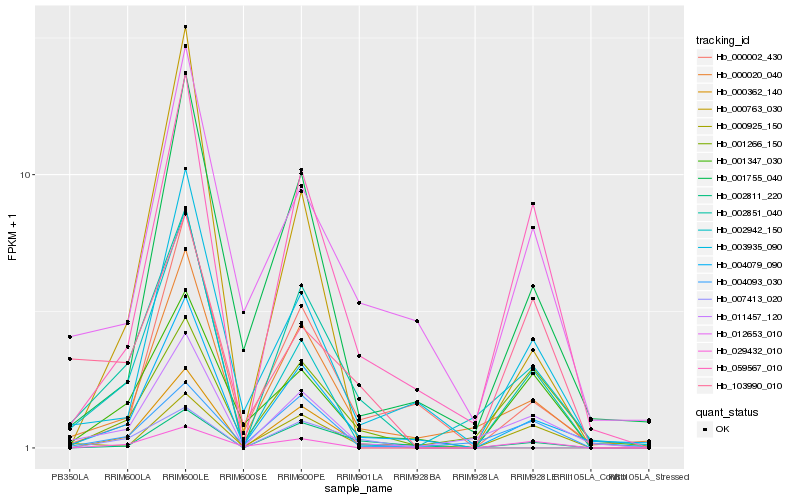
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002851_040 |
0.0 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 5 [Jatropha curcas] |
| 2 |
Hb_004079_090 |
0.1789692702 |
- |
- |
hypothetical protein RCOM_1053120 [Ricinus communis] |
| 3 |
Hb_011457_120 |
0.186377205 |
- |
- |
PREDICTED: sulfate transporter 3.1-like [Jatropha curcas] |
| 4 |
Hb_059567_010 |
0.1911344944 |
- |
- |
PREDICTED: transcription factor PAR2-like [Jatropha curcas] |
| 5 |
Hb_000020_040 |
0.1949363315 |
- |
- |
S-adenosylmethionine-dependent methyltransferase, putative [Ricinus communis] |
| 6 |
Hb_002811_220 |
0.2108390702 |
transcription factor |
TF Family: Trihelix |
transcription factor, putative [Ricinus communis] |
| 7 |
Hb_001347_030 |
0.2182976818 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 8 |
Hb_002942_150 |
0.2191348197 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 9 |
Hb_004093_030 |
0.2294091015 |
- |
- |
hypothetical protein RCOM_1406750 [Ricinus communis] |
| 10 |
Hb_000362_140 |
0.2320712084 |
transcription factor |
TF Family: bZIP |
PREDICTED: basic leucine zipper 61-like [Jatropha curcas] |
| 11 |
Hb_103990_010 |
0.2350523472 |
- |
- |
PREDICTED: light-induced protein, chloroplastic-like [Nicotiana sylvestris] |
| 12 |
Hb_000002_430 |
0.2352520411 |
- |
- |
PREDICTED: FHA domain-containing protein At4g14490 [Populus euphratica] |
| 13 |
Hb_029432_010 |
0.2373881889 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 14 |
Hb_003935_090 |
0.240453593 |
- |
- |
PREDICTED: uncharacterized protein At4g38062 [Jatropha curcas] |
| 15 |
Hb_000925_150 |
0.2405363608 |
- |
- |
PREDICTED: aluminum-activated malate transporter 8-like [Jatropha curcas] |
| 16 |
Hb_001266_150 |
0.2413379403 |
- |
- |
hypothetical protein JCGZ_14961 [Jatropha curcas] |
| 17 |
Hb_007413_020 |
0.2418538452 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001755_040 |
0.245280743 |
- |
- |
PREDICTED: cyclin-A2-4-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_000763_030 |
0.2495427676 |
- |
- |
Taxadien-5-alpha-ol O-acetyltransferase, putative [Ricinus communis] |
| 20 |
Hb_012653_010 |
0.2501026138 |
- |
- |
PREDICTED: subtilisin-like protease SBT2.5 [Jatropha curcas] |