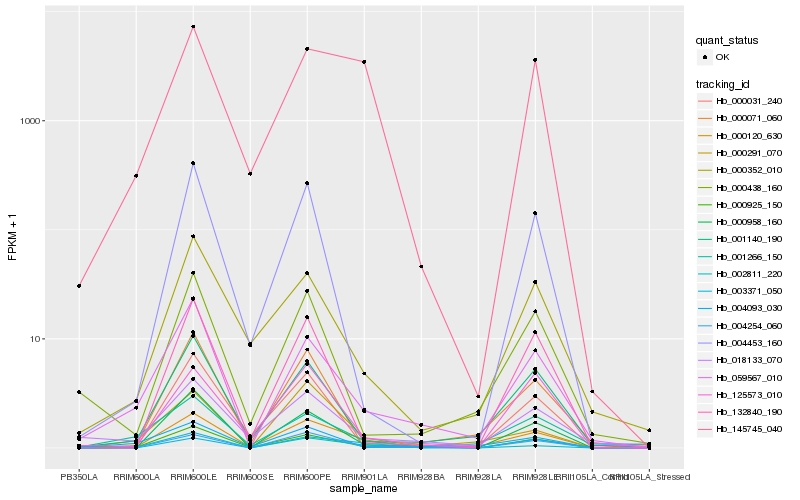
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000120_630 |
0.0 |
- |
- |
PREDICTED: glutamate receptor 3.6-like [Jatropha curcas] |
| 2 |
Hb_004254_060 |
0.1756936405 |
- |
- |
PREDICTED: uncharacterized protein LOC105650679 [Jatropha curcas] |
| 3 |
Hb_145745_040 |
0.1830060478 |
- |
- |
Cell wall-associated hydrolase [Medicago truncatula] |
| 4 |
Hb_001140_190 |
0.1862860395 |
- |
- |
PREDICTED: A-kinase anchor protein 9 [Jatropha curcas] |
| 5 |
Hb_000958_160 |
0.1915658108 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG1-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_002811_220 |
0.1950912842 |
transcription factor |
TF Family: Trihelix |
transcription factor, putative [Ricinus communis] |
| 7 |
Hb_000352_010 |
0.1951767685 |
- |
- |
PREDICTED: nudix hydrolase 13, mitochondrial [Jatropha curcas] |
| 8 |
Hb_059567_010 |
0.1957543247 |
- |
- |
PREDICTED: transcription factor PAR2-like [Jatropha curcas] |
| 9 |
Hb_003371_050 |
0.1963218368 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001266_150 |
0.2022453368 |
- |
- |
hypothetical protein JCGZ_14961 [Jatropha curcas] |
| 11 |
Hb_000071_060 |
0.206962109 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_004453_160 |
0.2073221096 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000291_070 |
0.2099598314 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_125573_010 |
0.210335581 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 15 |
Hb_132840_190 |
0.2114324874 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_000925_150 |
0.2116394535 |
- |
- |
PREDICTED: aluminum-activated malate transporter 8-like [Jatropha curcas] |
| 17 |
Hb_018133_070 |
0.2124436617 |
- |
- |
PREDICTED: RING-H2 finger protein ATL43 [Jatropha curcas] |
| 18 |
Hb_000031_240 |
0.213503018 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 19 |
Hb_004093_030 |
0.2136442814 |
- |
- |
hypothetical protein RCOM_1406750 [Ricinus communis] |
| 20 |
Hb_000438_160 |
0.2138443504 |
- |
- |
conserved hypothetical protein [Ricinus communis] |