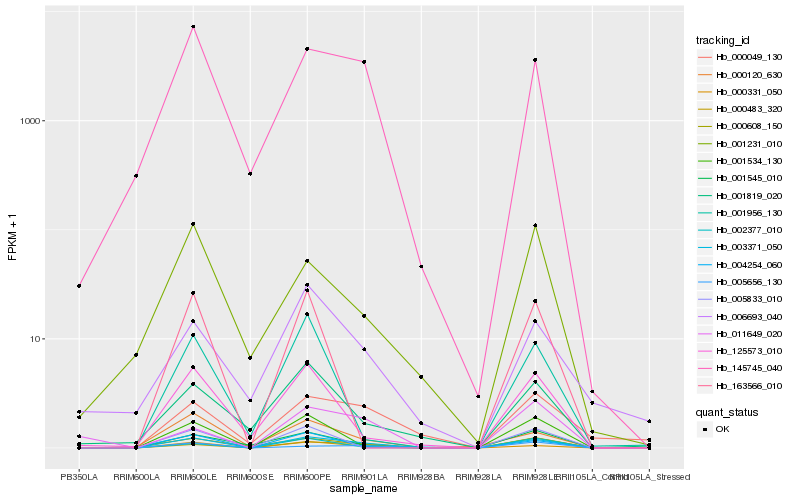
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003371_050 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000331_050 |
0.1559563845 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
hypothetical protein JCGZ_11303 [Jatropha curcas] |
| 3 |
Hb_145745_040 |
0.1817480184 |
- |
- |
Cell wall-associated hydrolase [Medicago truncatula] |
| 4 |
Hb_000120_630 |
0.1963218368 |
- |
- |
PREDICTED: glutamate receptor 3.6-like [Jatropha curcas] |
| 5 |
Hb_000608_150 |
0.2036645242 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 2 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000483_320 |
0.2144245821 |
- |
- |
Major pollen allergen Ory s 1 precursor, putative [Ricinus communis] |
| 7 |
Hb_001545_010 |
0.2152855703 |
- |
- |
PREDICTED: serine/threonine-protein kinase TNNI3K-like [Cucumis sativus] |
| 8 |
Hb_004254_060 |
0.2205264391 |
- |
- |
PREDICTED: uncharacterized protein LOC105650679 [Jatropha curcas] |
| 9 |
Hb_125573_010 |
0.2298460449 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 10 |
Hb_001534_130 |
0.2309357312 |
- |
- |
PREDICTED: serine carboxypeptidase-like 51 [Jatropha curcas] |
| 11 |
Hb_005656_130 |
0.2323303199 |
- |
- |
PREDICTED: calmodulin-like protein 7 [Jatropha curcas] |
| 12 |
Hb_002377_010 |
0.2376315993 |
transcription factor |
TF Family: C2C2-CO-like |
PREDICTED: zinc finger protein CONSTANS-LIKE 2-like [Jatropha curcas] |
| 13 |
Hb_001819_020 |
0.2394050559 |
- |
- |
PREDICTED: uncharacterized protein LOC105641890 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000049_130 |
0.2518337529 |
- |
- |
PREDICTED: uncharacterized protein LOC104879855 [Vitis vinifera] |
| 15 |
Hb_006693_040 |
0.2571552239 |
- |
- |
PREDICTED: homeotic protein female sterile-like [Jatropha curcas] |
| 16 |
Hb_001956_130 |
0.2584061884 |
- |
- |
catalytic, putative [Ricinus communis] |
| 17 |
Hb_163566_010 |
0.2670399534 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP17-1, chloroplastic isoform X2 [Amborella trichopoda] |
| 18 |
Hb_011649_020 |
0.2671208757 |
- |
- |
PREDICTED: uncharacterized protein LOC104212904 isoform X1 [Nicotiana sylvestris] |
| 19 |
Hb_001231_010 |
0.2689485767 |
- |
- |
calcium-binding family protein [Populus trichocarpa] |
| 20 |
Hb_005833_010 |
0.2690416747 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g46790, chloroplastic [Jatropha curcas] |