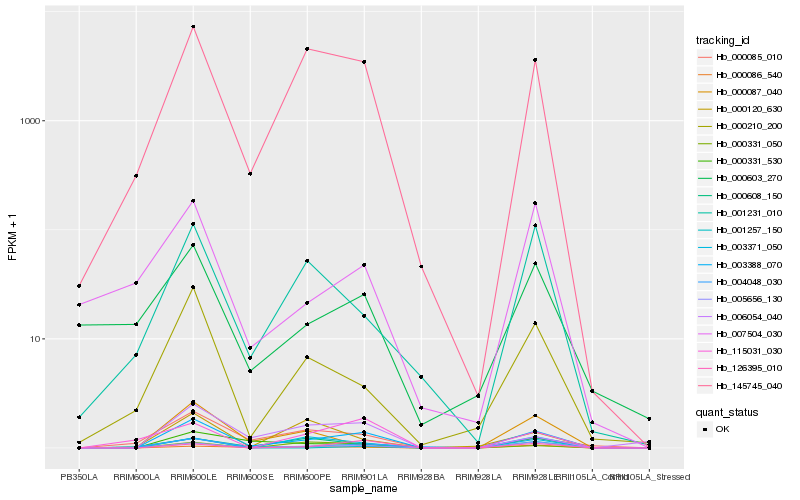
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003388_070 |
0.0 |
- |
- |
60S ribosomal protein L32A [Hevea brasiliensis] |
| 2 |
Hb_145745_040 |
0.1871473614 |
- |
- |
Cell wall-associated hydrolase [Medicago truncatula] |
| 3 |
Hb_005656_130 |
0.1926138137 |
- |
- |
PREDICTED: calmodulin-like protein 7 [Jatropha curcas] |
| 4 |
Hb_000331_530 |
0.2483791468 |
- |
- |
PREDICTED: callose synthase 10 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000210_200 |
0.2578928235 |
- |
- |
PREDICTED: exocyst complex component EXO70A1 [Jatropha curcas] |
| 6 |
Hb_006054_040 |
0.2583136221 |
- |
- |
- |
| 7 |
Hb_004048_030 |
0.2670695266 |
- |
- |
hypothetical protein B456_009G371200, partial [Gossypium raimondii] |
| 8 |
Hb_000086_540 |
0.2685335789 |
- |
- |
- |
| 9 |
Hb_126395_010 |
0.2717490668 |
- |
- |
hypothetical protein VITISV_043980 [Vitis vinifera] |
| 10 |
Hb_001231_010 |
0.2745763261 |
- |
- |
calcium-binding family protein [Populus trichocarpa] |
| 11 |
Hb_003371_050 |
0.274893391 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000085_010 |
0.2777963405 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 13 |
Hb_000331_050 |
0.2781459671 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
hypothetical protein JCGZ_11303 [Jatropha curcas] |
| 14 |
Hb_115031_030 |
0.2845222197 |
- |
- |
ribosomal protein S12 (chloroplast) [Camellia sinensis] |
| 15 |
Hb_000608_150 |
0.2921903617 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 2 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000120_630 |
0.2949614404 |
- |
- |
PREDICTED: glutamate receptor 3.6-like [Jatropha curcas] |
| 17 |
Hb_000603_270 |
0.3039179068 |
- |
- |
PREDICTED: uncharacterized protein LOC105638080 [Jatropha curcas] |
| 18 |
Hb_001257_150 |
0.304020885 |
- |
- |
- |
| 19 |
Hb_007504_030 |
0.3044748251 |
- |
- |
Rpl14, partial (chloroplast) [Schistostemon retusum] |
| 20 |
Hb_000087_040 |
0.3061959914 |
- |
- |
Autoinhibited Ca(2+)-ATPase 10 isoform 1 [Theobroma cacao] |