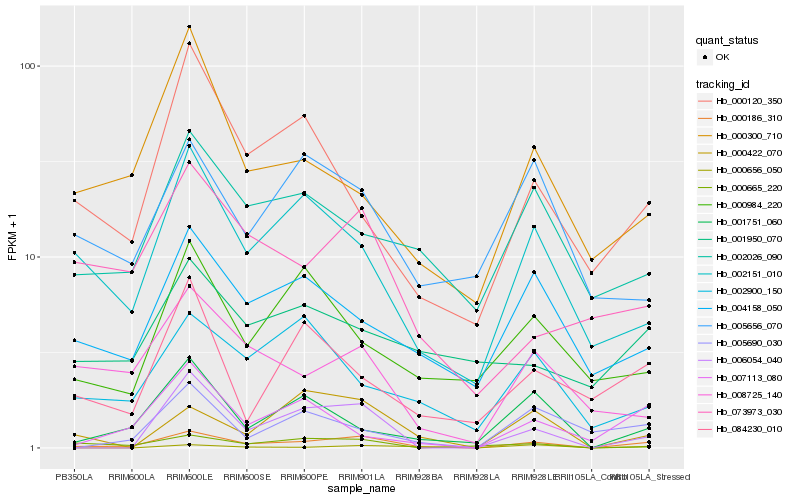
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000186_310 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_006054_040 |
0.2427354112 |
- |
- |
- |
| 3 |
Hb_002151_010 |
0.2567499814 |
- |
- |
hypothetical protein POPTR_0006s14610g [Populus trichocarpa] |
| 4 |
Hb_000422_070 |
0.2674322997 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_004158_050 |
0.270564692 |
- |
- |
hypothetical protein JCGZ_09026 [Jatropha curcas] |
| 6 |
Hb_008725_140 |
0.2731798637 |
- |
- |
PREDICTED: kynurenine--oxoglutarate transaminase-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_001751_060 |
0.2755020797 |
- |
- |
PREDICTED: SEC12-like protein 1 [Jatropha curcas] |
| 8 |
Hb_073973_030 |
0.2768149525 |
- |
- |
PREDICTED: uncharacterized protein LOC105638905 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000665_220 |
0.2800191303 |
- |
- |
catalytic, putative [Ricinus communis] |
| 10 |
Hb_000120_350 |
0.2800940929 |
- |
- |
PREDICTED: uncharacterized protein LOC104422872 isoform X1 [Eucalyptus grandis] |
| 11 |
Hb_002026_090 |
0.2819671924 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 4-like [Jatropha curcas] |
| 12 |
Hb_084230_010 |
0.2857548769 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 6 isoform X4 [Jatropha curcas] |
| 13 |
Hb_000656_050 |
0.2885689287 |
- |
- |
PREDICTED: 3-oxoacyl-[acyl-carrier-protein] synthase II, chloroplastic-like [Vitis vinifera] |
| 14 |
Hb_000300_710 |
0.2890136409 |
- |
- |
PREDICTED: uncharacterized protein LOC105123549 [Populus euphratica] |
| 15 |
Hb_007113_080 |
0.2903352594 |
- |
- |
PREDICTED: uncharacterized protein LOC105639553 isoform X1 [Jatropha curcas] |
| 16 |
Hb_002900_150 |
0.2922396979 |
- |
- |
PREDICTED: syntaxin-81 [Jatropha curcas] |
| 17 |
Hb_000984_220 |
0.293301045 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 2-like [Populus euphratica] |
| 18 |
Hb_005690_030 |
0.2957879365 |
- |
- |
hypothetical protein VITISV_020378 [Vitis vinifera] |
| 19 |
Hb_005656_070 |
0.2967447796 |
- |
- |
PREDICTED: cryptochrome DASH, chloroplastic/mitochondrial [Jatropha curcas] |
| 20 |
Hb_001950_070 |
0.2967793509 |
- |
- |
cyclin d, putative [Ricinus communis] |