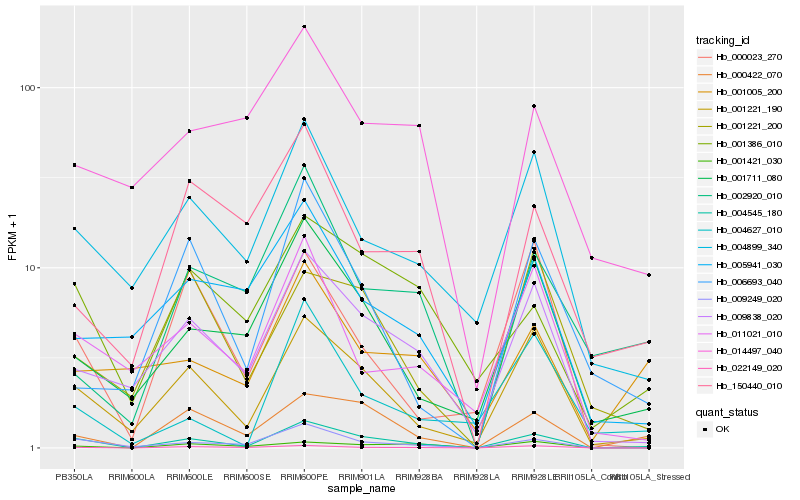
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004545_180 |
0.0 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 2 |
Hb_001221_190 |
0.1608502421 |
- |
- |
PREDICTED: F-box protein At5g50450 [Jatropha curcas] |
| 3 |
Hb_009249_020 |
0.1829460564 |
- |
- |
PREDICTED: endoglucanase 25-like [Jatropha curcas] |
| 4 |
Hb_000422_070 |
0.1931562 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_022149_020 |
0.2068733745 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_009838_020 |
0.2078899284 |
- |
- |
PREDICTED: cysteine-rich repeat secretory protein 3 [Jatropha curcas] |
| 7 |
Hb_001711_080 |
0.2221436472 |
transcription factor |
TF Family: C2C2-LSD |
PREDICTED: protein LSD1 isoform X1 [Phoenix dactylifera] |
| 8 |
Hb_004899_340 |
0.231721723 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 9 |
Hb_001386_010 |
0.2348209618 |
- |
- |
PREDICTED: uncharacterized protein LOC105632444 [Jatropha curcas] |
| 10 |
Hb_001421_030 |
0.2475712376 |
- |
- |
- |
| 11 |
Hb_011021_010 |
0.2554845112 |
- |
- |
kinase, putative [Ricinus communis] |
| 12 |
Hb_004627_010 |
0.2656043419 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_005941_030 |
0.2657420182 |
transcription factor |
TF Family: HB |
DNA binding protein, putative [Ricinus communis] |
| 14 |
Hb_001221_200 |
0.2659630502 |
- |
- |
- |
| 15 |
Hb_006693_040 |
0.2663865479 |
- |
- |
PREDICTED: homeotic protein female sterile-like [Jatropha curcas] |
| 16 |
Hb_000023_270 |
0.267851708 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002920_010 |
0.2685963262 |
- |
- |
PREDICTED: sulfiredoxin, chloroplastic/mitochondrial [Populus euphratica] |
| 18 |
Hb_150440_010 |
0.2695356174 |
- |
- |
- |
| 19 |
Hb_001005_200 |
0.2742728425 |
- |
- |
PREDICTED: receptor-like protein kinase HERK 1 isoform X2 [Jatropha curcas] |
| 20 |
Hb_014497_040 |
0.2767955447 |
- |
- |
PREDICTED: calvin cycle protein CP12-3, chloroplastic [Jatropha curcas] |