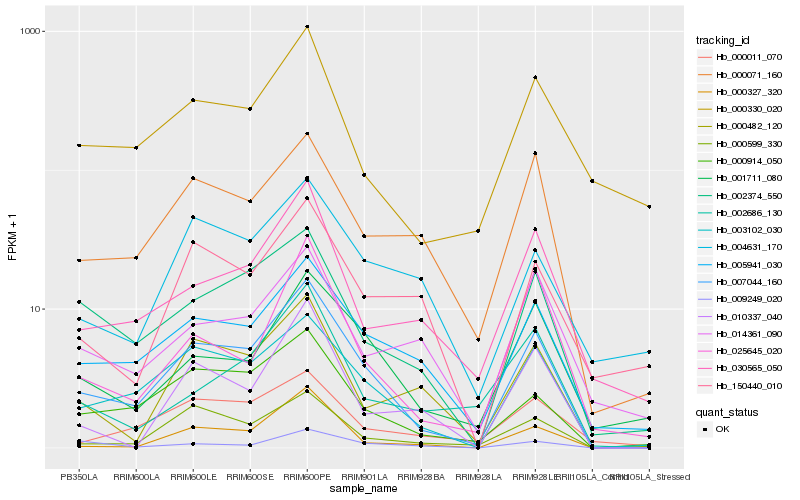
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007044_160 |
0.0 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor CYCLOIDEA [Populus euphratica] |
| 2 |
Hb_003102_030 |
0.121540783 |
- |
- |
PREDICTED: uncharacterized protein LOC105644418 [Jatropha curcas] |
| 3 |
Hb_000914_050 |
0.1502851866 |
- |
- |
PREDICTED: serine/threonine-protein kinase UCNL [Jatropha curcas] |
| 4 |
Hb_005941_030 |
0.1518078644 |
transcription factor |
TF Family: HB |
DNA binding protein, putative [Ricinus communis] |
| 5 |
Hb_002374_550 |
0.1649123007 |
- |
- |
PREDICTED: aspartic proteinase nepenthesin-1 [Jatropha curcas] |
| 6 |
Hb_000071_160 |
0.1751609375 |
- |
- |
PREDICTED: protein IQ-DOMAIN 14 [Jatropha curcas] |
| 7 |
Hb_001711_080 |
0.1768570583 |
transcription factor |
TF Family: C2C2-LSD |
PREDICTED: protein LSD1 isoform X1 [Phoenix dactylifera] |
| 8 |
Hb_000599_330 |
0.1772684598 |
- |
- |
hypothetical protein JCGZ_17661 [Jatropha curcas] |
| 9 |
Hb_000482_120 |
0.1824983961 |
- |
- |
PREDICTED: uncharacterized protein LOC105638146 [Jatropha curcas] |
| 10 |
Hb_030565_050 |
0.1859102315 |
- |
- |
UDP-D-GLUCURONATE 4-EPIMERASE 6 family protein [Populus trichocarpa] |
| 11 |
Hb_025645_020 |
0.1887050434 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 12 |
Hb_000327_320 |
0.1888751554 |
- |
- |
PREDICTED: probable receptor-like protein kinase At2g39360 [Jatropha curcas] |
| 13 |
Hb_000011_070 |
0.1927178611 |
- |
- |
PREDICTED: CO(2)-response secreted protease-like [Jatropha curcas] |
| 14 |
Hb_010337_040 |
0.1954442316 |
- |
- |
- |
| 15 |
Hb_002686_130 |
0.2005560934 |
transcription factor |
TF Family: G2-like |
PREDICTED: uncharacterized protein LOC105650401 [Jatropha curcas] |
| 16 |
Hb_000330_020 |
0.2016027758 |
- |
- |
PREDICTED: isoflavone reductase-like protein [Jatropha curcas] |
| 17 |
Hb_009249_020 |
0.2060877132 |
- |
- |
PREDICTED: endoglucanase 25-like [Jatropha curcas] |
| 18 |
Hb_004631_170 |
0.2081716261 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_014361_090 |
0.2096114025 |
- |
- |
acyl-CoA binding protein 3A [Vernicia fordii] |
| 20 |
Hb_150440_010 |
0.2099888352 |
- |
- |
- |