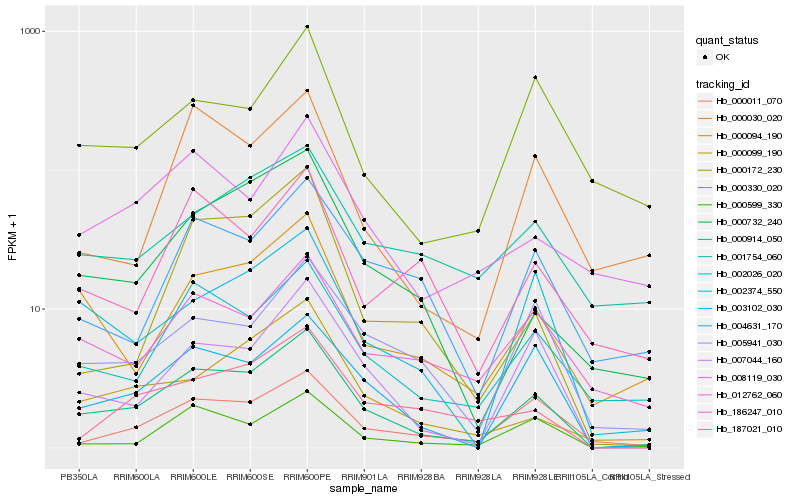
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000914_050 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein kinase UCNL [Jatropha curcas] |
| 2 |
Hb_003102_030 |
0.1415385184 |
- |
- |
PREDICTED: uncharacterized protein LOC105644418 [Jatropha curcas] |
| 3 |
Hb_007044_160 |
0.1502851866 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor CYCLOIDEA [Populus euphratica] |
| 4 |
Hb_002026_020 |
0.1569904338 |
- |
- |
hypothetical protein POPTR_0012s129501g, partial [Populus trichocarpa] |
| 5 |
Hb_002374_550 |
0.1702328651 |
- |
- |
PREDICTED: aspartic proteinase nepenthesin-1 [Jatropha curcas] |
| 6 |
Hb_000094_190 |
0.1705898631 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 7-like [Jatropha curcas] |
| 7 |
Hb_005941_030 |
0.1714964474 |
transcription factor |
TF Family: HB |
DNA binding protein, putative [Ricinus communis] |
| 8 |
Hb_000011_070 |
0.1716900819 |
- |
- |
PREDICTED: CO(2)-response secreted protease-like [Jatropha curcas] |
| 9 |
Hb_000599_330 |
0.1737375194 |
- |
- |
hypothetical protein JCGZ_17661 [Jatropha curcas] |
| 10 |
Hb_000732_240 |
0.1747169789 |
- |
- |
PREDICTED: dynein light chain 1, cytoplasmic-like [Jatropha curcas] |
| 11 |
Hb_000099_190 |
0.1761930174 |
- |
- |
PREDICTED: putative cell division cycle ATPase [Jatropha curcas] |
| 12 |
Hb_012762_060 |
0.1863443127 |
- |
- |
Vitellogenic carboxypeptidase, putative [Ricinus communis] |
| 13 |
Hb_004631_170 |
0.1886976395 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_187021_010 |
0.1887993246 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 15 |
Hb_008119_030 |
0.191778869 |
- |
- |
PREDICTED: peroxidase 63-like [Jatropha curcas] |
| 16 |
Hb_000030_020 |
0.1974818093 |
- |
- |
PREDICTED: uncharacterized protein LOC105123549 [Populus euphratica] |
| 17 |
Hb_000172_230 |
0.1998566628 |
- |
- |
myo-inositol-1 phosphate synthase [Hevea brasiliensis] |
| 18 |
Hb_000330_020 |
0.2005373053 |
- |
- |
PREDICTED: isoflavone reductase-like protein [Jatropha curcas] |
| 19 |
Hb_001754_060 |
0.2016799317 |
- |
- |
Xyloglucan 6-xylosyltransferase, putative [Ricinus communis] |
| 20 |
Hb_186247_010 |
0.2028588985 |
- |
- |
PREDICTED: uridine 5'-monophosphate synthase-like [Pyrus x bretschneideri] |