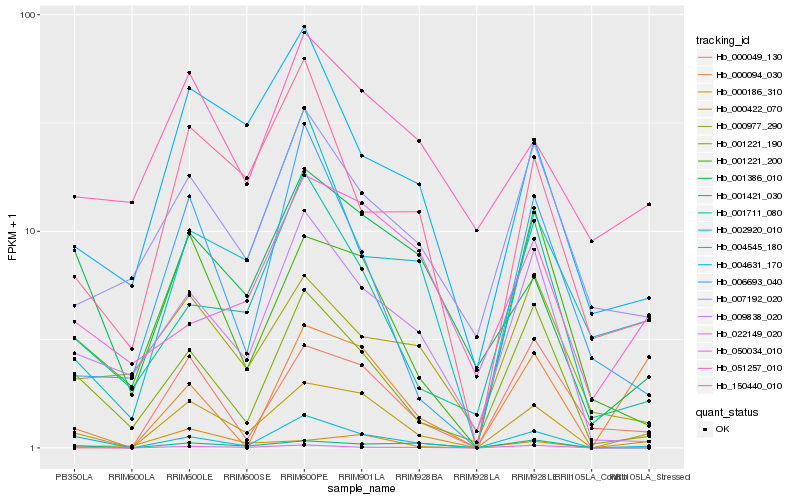
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000422_070 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_004545_180 |
0.1931562 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 3 |
Hb_001221_190 |
0.2258154815 |
- |
- |
PREDICTED: F-box protein At5g50450 [Jatropha curcas] |
| 4 |
Hb_001221_200 |
0.2317659229 |
- |
- |
- |
| 5 |
Hb_009838_020 |
0.2397851476 |
- |
- |
PREDICTED: cysteine-rich repeat secretory protein 3 [Jatropha curcas] |
| 6 |
Hb_001386_010 |
0.240255572 |
- |
- |
PREDICTED: uncharacterized protein LOC105632444 [Jatropha curcas] |
| 7 |
Hb_000094_030 |
0.2496713348 |
- |
- |
- |
| 8 |
Hb_001711_080 |
0.2504683683 |
transcription factor |
TF Family: C2C2-LSD |
PREDICTED: protein LSD1 isoform X1 [Phoenix dactylifera] |
| 9 |
Hb_000049_130 |
0.252416703 |
- |
- |
PREDICTED: uncharacterized protein LOC104879855 [Vitis vinifera] |
| 10 |
Hb_150440_010 |
0.2530165549 |
- |
- |
- |
| 11 |
Hb_007192_020 |
0.2548383636 |
- |
- |
PREDICTED: high-affinity nitrate transporter 3.1-like [Jatropha curcas] |
| 12 |
Hb_004631_170 |
0.2596069855 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_022149_020 |
0.2598156917 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001421_030 |
0.2606467767 |
- |
- |
- |
| 15 |
Hb_051257_010 |
0.2617567315 |
- |
- |
- |
| 16 |
Hb_002920_010 |
0.2651728234 |
- |
- |
PREDICTED: sulfiredoxin, chloroplastic/mitochondrial [Populus euphratica] |
| 17 |
Hb_000186_310 |
0.2674322997 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_006693_040 |
0.2677109813 |
- |
- |
PREDICTED: homeotic protein female sterile-like [Jatropha curcas] |
| 19 |
Hb_050034_010 |
0.271097838 |
- |
- |
PREDICTED: cullin-1 [Jatropha curcas] |
| 20 |
Hb_000977_290 |
0.2752334044 |
- |
- |
PREDICTED: CMP-sialic acid transporter 2-like isoform X2 [Jatropha curcas] |