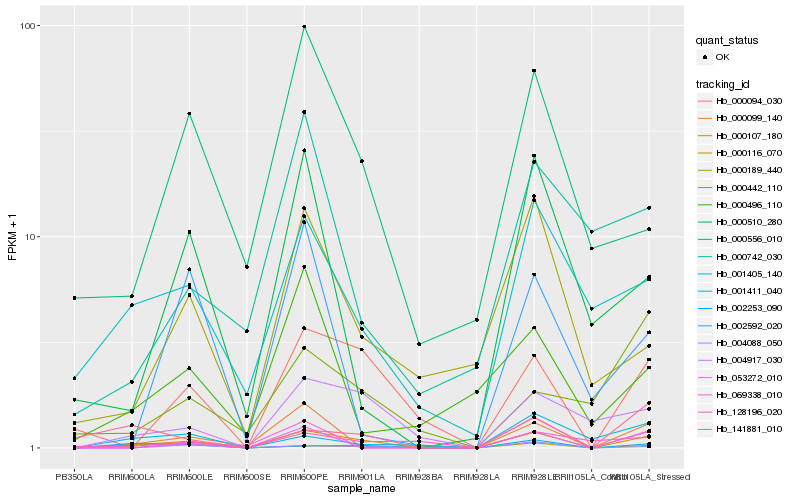
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000107_180 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103343662 [Prunus mume] |
| 2 |
Hb_000094_030 |
0.2564597231 |
- |
- |
- |
| 3 |
Hb_004917_030 |
0.2715222571 |
- |
- |
- |
| 4 |
Hb_002253_090 |
0.288376018 |
- |
- |
Chitinase 1 precursor, putative [Ricinus communis] |
| 5 |
Hb_001405_140 |
0.294382802 |
- |
- |
hypothetical protein POPTR_0004s08350g, partial [Populus trichocarpa] |
| 6 |
Hb_000099_140 |
0.2987073949 |
- |
- |
PREDICTED: aluminum-activated malate transporter 9-like [Jatropha curcas] |
| 7 |
Hb_004088_050 |
0.3027168255 |
- |
- |
hypothetical protein VITISV_027174 [Vitis vinifera] |
| 8 |
Hb_053272_010 |
0.312399039 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1-like [Jatropha curcas] |
| 9 |
Hb_000116_070 |
0.3137485522 |
transcription factor |
TF Family: C2H2 |
hypothetical protein JCGZ_22381 [Jatropha curcas] |
| 10 |
Hb_000556_010 |
0.3149765356 |
- |
- |
glycerate dehydrogenase, putative [Ricinus communis] |
| 11 |
Hb_000189_440 |
0.3209835473 |
- |
- |
hypothetical protein JCGZ_23470 [Jatropha curcas] |
| 12 |
Hb_141881_010 |
0.3253988893 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 13 |
Hb_000510_280 |
0.3299428833 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-induced protein 22D-like [Jatropha curcas] |
| 14 |
Hb_069338_010 |
0.330597216 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001411_040 |
0.3368376195 |
- |
- |
hypothetical protein VITISV_010644 [Vitis vinifera] |
| 16 |
Hb_128196_020 |
0.3397916599 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 3, chloroplastic [Nelumbo nucifera] |
| 17 |
Hb_002592_020 |
0.3447243834 |
- |
- |
PREDICTED: probable tyrosine-protein phosphatase At1g05000 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000496_110 |
0.3474963296 |
- |
- |
PREDICTED: uncharacterized protein LOC105650214 [Jatropha curcas] |
| 19 |
Hb_000742_030 |
0.3508956136 |
- |
- |
PREDICTED: uncharacterized protein LOC105636851 [Jatropha curcas] |
| 20 |
Hb_000442_110 |
0.3510216554 |
- |
- |
hypothetical protein POPTR_0010s18960g [Populus trichocarpa] |