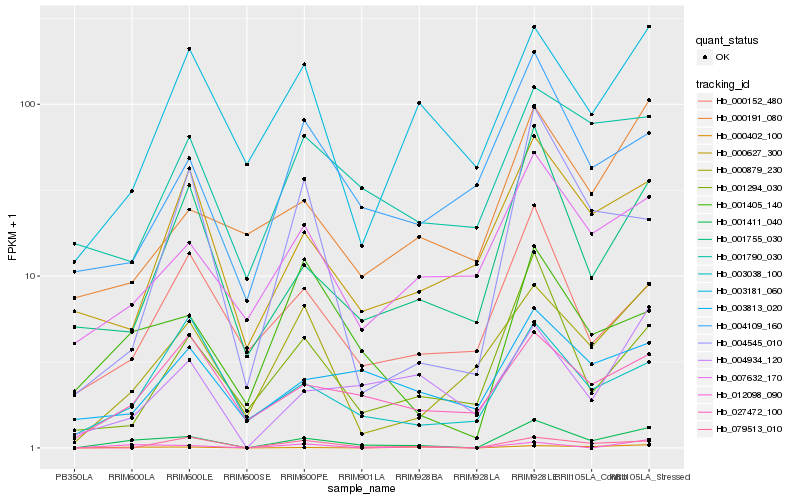
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001411_040 |
0.0 |
- |
- |
hypothetical protein VITISV_010644 [Vitis vinifera] |
| 2 |
Hb_001755_030 |
0.2409563218 |
- |
- |
PREDICTED: D-glycerate 3-kinase, chloroplastic-like [Populus euphratica] |
| 3 |
Hb_000402_100 |
0.2437622955 |
- |
- |
PREDICTED: L-tryptophan--pyruvate aminotransferase 1 [Jatropha curcas] |
| 4 |
Hb_001405_140 |
0.2533736568 |
- |
- |
hypothetical protein POPTR_0004s08350g, partial [Populus trichocarpa] |
| 5 |
Hb_000879_230 |
0.254807289 |
- |
- |
PREDICTED: tetratricopeptide repeat protein 7A [Jatropha curcas] |
| 6 |
Hb_001294_030 |
0.2619565528 |
- |
- |
PREDICTED: uncharacterized protein LOC105644307 [Jatropha curcas] |
| 7 |
Hb_007632_170 |
0.2624866075 |
- |
- |
hypothetical protein JCGZ_01028 [Jatropha curcas] |
| 8 |
Hb_004934_120 |
0.2625830985 |
- |
- |
- |
| 9 |
Hb_027472_100 |
0.2640311763 |
- |
- |
PREDICTED: uncharacterized protein LOC105646173 [Jatropha curcas] |
| 10 |
Hb_000627_300 |
0.2712720186 |
- |
- |
PREDICTED: uncharacterized protein LOC105632672 [Jatropha curcas] |
| 11 |
Hb_004545_010 |
0.2740111404 |
- |
- |
acyl carrier protein [Ricinus communis] |
| 12 |
Hb_079513_010 |
0.2752243506 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000152_480 |
0.2753421396 |
- |
- |
PREDICTED: protein LOW PSII ACCUMULATION 2, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000191_080 |
0.2762695353 |
- |
- |
PREDICTED: uncharacterized protein LOC105637945 [Jatropha curcas] |
| 15 |
Hb_003181_060 |
0.2764197071 |
- |
- |
PREDICTED: triosephosphate isomerase, cytosolic [Jatropha curcas] |
| 16 |
Hb_003038_100 |
0.2766183636 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |
| 17 |
Hb_012098_090 |
0.2779496447 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 18 |
Hb_003813_020 |
0.2785527432 |
- |
- |
PREDICTED: uncharacterized protein LOC105638736 isoform X1 [Jatropha curcas] |
| 19 |
Hb_004109_160 |
0.2798176053 |
- |
- |
PREDICTED: peptide methionine sulfoxide reductase B1, chloroplastic [Jatropha curcas] |
| 20 |
Hb_001790_030 |
0.280198294 |
- |
- |
PREDICTED: ferredoxin-thioredoxin reductase catalytic chain, chloroplastic [Jatropha curcas] |