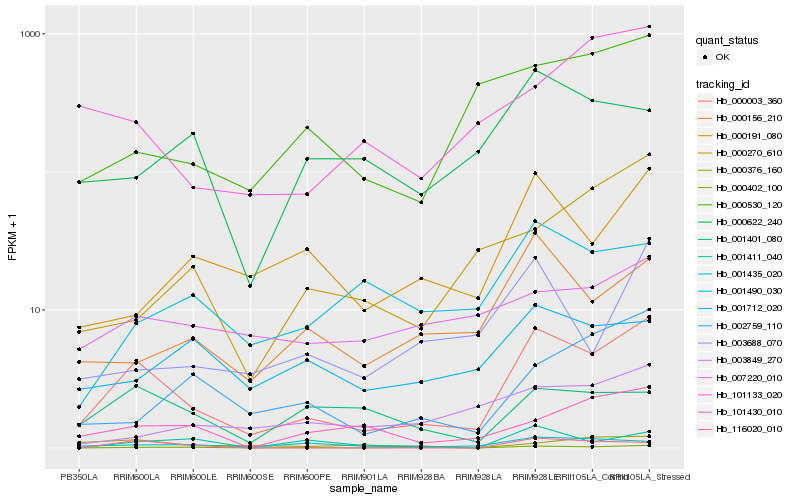
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000003_360 |
0.0 |
- |
- |
Putative inactive purple acid phosphatase 27 [Glycine soja] |
| 2 |
Hb_000402_100 |
0.2478417119 |
- |
- |
PREDICTED: L-tryptophan--pyruvate aminotransferase 1 [Jatropha curcas] |
| 3 |
Hb_000156_210 |
0.2580256999 |
- |
- |
PREDICTED: metal transporter Nramp1 [Jatropha curcas] |
| 4 |
Hb_000191_080 |
0.2614083852 |
- |
- |
PREDICTED: uncharacterized protein LOC105637945 [Jatropha curcas] |
| 5 |
Hb_001435_020 |
0.268959888 |
- |
- |
hypothetical protein JCGZ_22466 [Jatropha curcas] |
| 6 |
Hb_101430_010 |
0.2700110806 |
- |
- |
- |
| 7 |
Hb_000530_120 |
0.2707101468 |
- |
- |
copper transport protein ATOX1 [Hevea brasiliensis] |
| 8 |
Hb_002759_110 |
0.2738706657 |
- |
- |
RNA-binding family protein, putative isoform 1 [Theobroma cacao] |
| 9 |
Hb_101133_020 |
0.2748626025 |
- |
- |
PREDICTED: 15.7 kDa heat shock protein, peroxisomal [Jatropha curcas] |
| 10 |
Hb_003688_070 |
0.2752036081 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein E [Jatropha curcas] |
| 11 |
Hb_001401_080 |
0.2858788734 |
- |
- |
PREDICTED: protein TIC 22, chloroplastic [Jatropha curcas] |
| 12 |
Hb_001490_030 |
0.287403485 |
- |
- |
PREDICTED: psbP domain-containing protein 6, chloroplastic [Jatropha curcas] |
| 13 |
Hb_001411_040 |
0.2876816083 |
- |
- |
hypothetical protein VITISV_010644 [Vitis vinifera] |
| 14 |
Hb_000270_610 |
0.2893751815 |
- |
- |
PREDICTED: uncharacterized protein At5g50100, mitochondrial [Jatropha curcas] |
| 15 |
Hb_007220_010 |
0.2919891086 |
- |
- |
PREDICTED: methylcrotonoyl-CoA carboxylase beta chain, mitochondrial [Jatropha curcas] |
| 16 |
Hb_001712_020 |
0.2939909343 |
- |
- |
PREDICTED: uncharacterized protein LOC105641948 [Jatropha curcas] |
| 17 |
Hb_116020_010 |
0.297181568 |
- |
- |
Uncharacterized protein TCM_023639 [Theobroma cacao] |
| 18 |
Hb_000622_240 |
0.2980397739 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1A, chloroplastic [Jatropha curcas] |
| 19 |
Hb_003849_270 |
0.2980469994 |
- |
- |
- |
| 20 |
Hb_000376_160 |
0.2987396348 |
- |
- |
- |