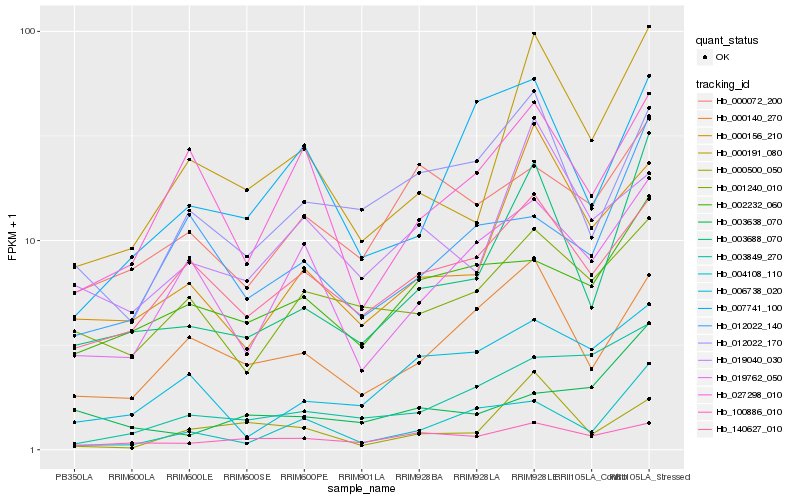
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003688_070 |
0.0 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein E [Jatropha curcas] |
| 2 |
Hb_000191_080 |
0.1468280977 |
- |
- |
PREDICTED: uncharacterized protein LOC105637945 [Jatropha curcas] |
| 3 |
Hb_000156_210 |
0.171841585 |
- |
- |
PREDICTED: metal transporter Nramp1 [Jatropha curcas] |
| 4 |
Hb_004108_110 |
0.1809014828 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000140_270 |
0.198206446 |
- |
- |
S-adenosyl-L-methionine-dependent methyltransferases superfamily protein [Theobroma cacao] |
| 6 |
Hb_012022_170 |
0.2009564387 |
transcription factor |
TF Family: HMG |
DNA-binding protein MNB1B, putative [Ricinus communis] |
| 7 |
Hb_140627_010 |
0.2172807504 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_007741_100 |
0.2179254994 |
transcription factor |
TF Family: GRAS |
PREDICTED: LOW QUALITY PROTEIN: scarecrow-like protein 6 [Jatropha curcas] |
| 9 |
Hb_012022_140 |
0.2187839146 |
- |
- |
ADP,ATP carrier protein, putative [Ricinus communis] |
| 10 |
Hb_019762_050 |
0.2190962541 |
transcription factor |
TF Family: CSD |
PREDICTED: glycine-rich protein 2-like [Jatropha curcas] |
| 11 |
Hb_006738_020 |
0.22612301 |
- |
- |
hypothetical protein VITISV_010510 [Vitis vinifera] |
| 12 |
Hb_001240_010 |
0.2269291353 |
- |
- |
PREDICTED: uncharacterized protein LOC105634101 [Jatropha curcas] |
| 13 |
Hb_002232_060 |
0.2292777046 |
- |
- |
PREDICTED: dentin sialophosphoprotein [Jatropha curcas] |
| 14 |
Hb_003849_270 |
0.2305209688 |
- |
- |
- |
| 15 |
Hb_100886_010 |
0.2308786603 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 16 |
Hb_027298_010 |
0.2320308096 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_003638_070 |
0.2327913451 |
- |
- |
hypothetical protein EUGRSUZ_E00638 [Eucalyptus grandis] |
| 18 |
Hb_019040_030 |
0.233858017 |
- |
- |
PREDICTED: probable ribosome-binding factor A, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000072_200 |
0.2351762103 |
- |
- |
PREDICTED: myb-like protein X [Jatropha curcas] |
| 20 |
Hb_000500_050 |
0.2368274787 |
- |
- |
PREDICTED: uncharacterized protein LOC105160092 [Sesamum indicum] |