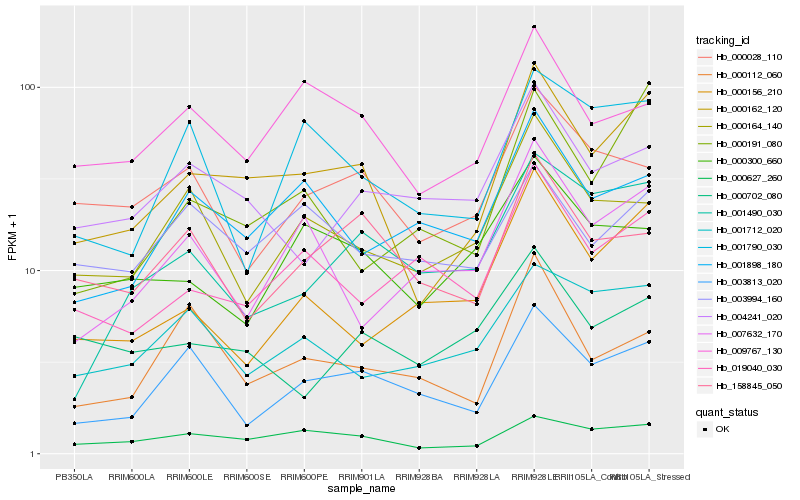
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000162_120 |
0.0 |
- |
- |
PREDICTED: chaperone protein dnaJ 11, chloroplastic-like [Jatropha curcas] |
| 2 |
Hb_000627_260 |
0.1421942176 |
- |
- |
chloride channel clc, putative [Ricinus communis] |
| 3 |
Hb_000191_080 |
0.1703415085 |
- |
- |
PREDICTED: uncharacterized protein LOC105637945 [Jatropha curcas] |
| 4 |
Hb_009767_130 |
0.1718013017 |
- |
- |
PREDICTED: preprotein translocase subunit SECE1 [Jatropha curcas] |
| 5 |
Hb_003813_020 |
0.1803491463 |
- |
- |
PREDICTED: uncharacterized protein LOC105638736 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001898_180 |
0.1865300916 |
- |
- |
PREDICTED: translation initiation factor IF-1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_019040_030 |
0.1880827518 |
- |
- |
PREDICTED: probable ribosome-binding factor A, chloroplastic [Jatropha curcas] |
| 8 |
Hb_001712_020 |
0.1888000392 |
- |
- |
PREDICTED: uncharacterized protein LOC105641948 [Jatropha curcas] |
| 9 |
Hb_001490_030 |
0.1891911861 |
- |
- |
PREDICTED: psbP domain-containing protein 6, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000156_210 |
0.1906353479 |
- |
- |
PREDICTED: metal transporter Nramp1 [Jatropha curcas] |
| 11 |
Hb_004241_020 |
0.1913071655 |
- |
- |
PREDICTED: DAG protein, chloroplastic-like [Jatropha curcas] |
| 12 |
Hb_000164_140 |
0.1921080289 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 13 |
Hb_003994_160 |
0.1934002771 |
- |
- |
PREDICTED: uncharacterized protein LOC105131691 isoform X5 [Populus euphratica] |
| 14 |
Hb_007632_170 |
0.1937420146 |
- |
- |
hypothetical protein JCGZ_01028 [Jatropha curcas] |
| 15 |
Hb_001790_030 |
0.1950394246 |
- |
- |
PREDICTED: ferredoxin-thioredoxin reductase catalytic chain, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000028_110 |
0.1952825423 |
- |
- |
PREDICTED: uncharacterized protein LOC105645305 [Jatropha curcas] |
| 17 |
Hb_158845_050 |
0.1953087215 |
- |
- |
PREDICTED: uncharacterized protein LOC105122890 [Populus euphratica] |
| 18 |
Hb_000300_660 |
0.1970334874 |
- |
- |
unknown [Glycine max] |
| 19 |
Hb_000112_060 |
0.1993292069 |
- |
- |
PREDICTED: thioredoxin-like protein CITRX, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000702_080 |
0.200773263 |
- |
- |
PREDICTED: uncharacterized protein LOC105630492 [Jatropha curcas] |