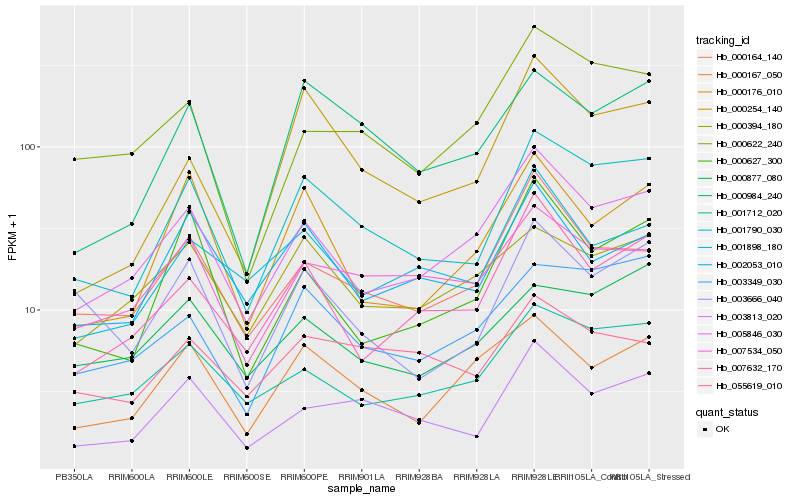
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001790_030 |
0.0 |
- |
- |
PREDICTED: ferredoxin-thioredoxin reductase catalytic chain, chloroplastic [Jatropha curcas] |
| 2 |
Hb_003813_020 |
0.1138370844 |
- |
- |
PREDICTED: uncharacterized protein LOC105638736 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000984_240 |
0.1169664607 |
- |
- |
hypothetical protein JCGZ_15962 [Jatropha curcas] |
| 4 |
Hb_003349_030 |
0.120150764 |
- |
- |
PREDICTED: uncharacterized protein LOC105628283 [Jatropha curcas] |
| 5 |
Hb_005846_030 |
0.1261533331 |
- |
- |
PREDICTED: 50S ribosomal protein L29, chloroplastic [Jatropha curcas] |
| 6 |
Hb_001712_020 |
0.130278359 |
- |
- |
PREDICTED: uncharacterized protein LOC105641948 [Jatropha curcas] |
| 7 |
Hb_000254_140 |
0.1324800086 |
- |
- |
PREDICTED: 30S ribosomal protein S17, chloroplastic [Jatropha curcas] |
| 8 |
Hb_003666_040 |
0.1325549018 |
- |
- |
PREDICTED: uncharacterized protein LOC105639624 [Jatropha curcas] |
| 9 |
Hb_000164_140 |
0.141041941 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 10 |
Hb_000176_010 |
0.1413977309 |
- |
- |
PREDICTED: 50S ribosomal protein L35, chloroplastic [Jatropha curcas] |
| 11 |
Hb_007534_050 |
0.1427502022 |
- |
- |
PREDICTED: glutamyl-tRNA reductase-binding protein, chloroplastic isoform X1 [Jatropha curcas] |
| 12 |
Hb_007632_170 |
0.1427610252 |
- |
- |
hypothetical protein JCGZ_01028 [Jatropha curcas] |
| 13 |
Hb_000167_050 |
0.1439156697 |
- |
- |
PREDICTED: uncharacterized protein LOC105641106 [Jatropha curcas] |
| 14 |
Hb_000622_240 |
0.143924148 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1A, chloroplastic [Jatropha curcas] |
| 15 |
Hb_002053_010 |
0.1459954115 |
- |
- |
PREDICTED: clp protease-related protein At4g12060, chloroplastic-like [Jatropha curcas] |
| 16 |
Hb_000877_080 |
0.1469772367 |
- |
- |
PREDICTED: uncharacterized protein LOC105640901 [Jatropha curcas] |
| 17 |
Hb_000627_300 |
0.1474207363 |
- |
- |
PREDICTED: uncharacterized protein LOC105632672 [Jatropha curcas] |
| 18 |
Hb_001898_180 |
0.1496184384 |
- |
- |
PREDICTED: translation initiation factor IF-1, chloroplastic [Jatropha curcas] |
| 19 |
Hb_055619_010 |
0.1511841877 |
- |
- |
PREDICTED: uncharacterized protein LOC105632456 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000394_180 |
0.152495437 |
- |
- |
PREDICTED: uncharacterized protein LOC105636995 [Jatropha curcas] |