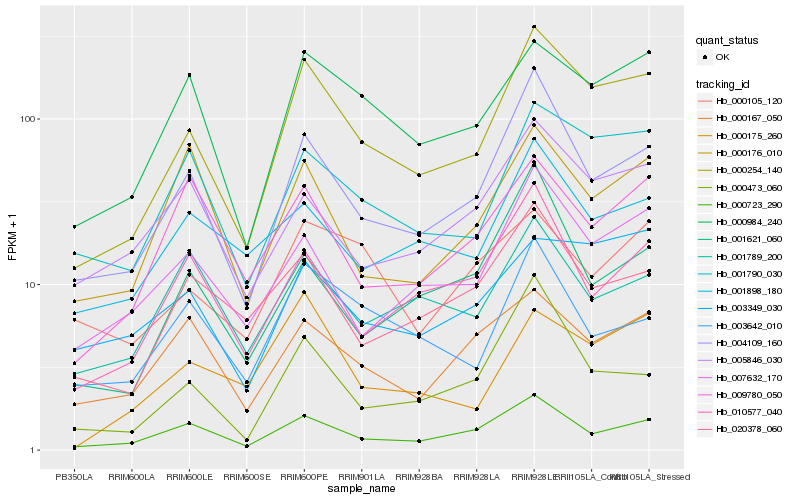
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000254_140 |
0.0 |
- |
- |
PREDICTED: 30S ribosomal protein S17, chloroplastic [Jatropha curcas] |
| 2 |
Hb_004109_160 |
0.116167106 |
- |
- |
PREDICTED: peptide methionine sulfoxide reductase B1, chloroplastic [Jatropha curcas] |
| 3 |
Hb_001790_030 |
0.1324800086 |
- |
- |
PREDICTED: ferredoxin-thioredoxin reductase catalytic chain, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000984_240 |
0.1343574403 |
- |
- |
hypothetical protein JCGZ_15962 [Jatropha curcas] |
| 5 |
Hb_000723_290 |
0.1365402571 |
- |
- |
calcium lipid binding protein, putative [Ricinus communis] |
| 6 |
Hb_007632_170 |
0.1472196504 |
- |
- |
hypothetical protein JCGZ_01028 [Jatropha curcas] |
| 7 |
Hb_020378_060 |
0.1645229166 |
- |
- |
PREDICTED: methyltransferase-like protein 13 [Jatropha curcas] |
| 8 |
Hb_000175_260 |
0.1662907376 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 14-like [Populus euphratica] |
| 9 |
Hb_000473_060 |
0.1674697757 |
- |
- |
PREDICTED: psbP domain-containing protein 2, chloroplastic [Jatropha curcas] |
| 10 |
Hb_003349_030 |
0.1682744253 |
- |
- |
PREDICTED: uncharacterized protein LOC105628283 [Jatropha curcas] |
| 11 |
Hb_001898_180 |
0.168758326 |
- |
- |
PREDICTED: translation initiation factor IF-1, chloroplastic [Jatropha curcas] |
| 12 |
Hb_010577_040 |
0.1688229845 |
- |
- |
PREDICTED: uncharacterized protein LOC105643738 [Jatropha curcas] |
| 13 |
Hb_005846_030 |
0.1697220184 |
- |
- |
PREDICTED: 50S ribosomal protein L29, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000167_050 |
0.1752523989 |
- |
- |
PREDICTED: uncharacterized protein LOC105641106 [Jatropha curcas] |
| 15 |
Hb_003642_010 |
0.1763818171 |
- |
- |
PREDICTED: chlorophyll(ide) b reductase NOL, chloroplastic isoform X2 [Jatropha curcas] |
| 16 |
Hb_000176_010 |
0.1822785587 |
- |
- |
PREDICTED: 50S ribosomal protein L35, chloroplastic [Jatropha curcas] |
| 17 |
Hb_009780_050 |
0.1829196491 |
- |
- |
PREDICTED: glutathione S-transferase DHAR3, chloroplastic [Jatropha curcas] |
| 18 |
Hb_001789_200 |
0.1838260318 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATB, chloroplastic [Jatropha curcas] |
| 19 |
Hb_001621_060 |
0.1840741608 |
- |
- |
PREDICTED: thylakoid lumenal 15 kDa protein 1, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000105_120 |
0.1841671005 |
- |
- |
PREDICTED: uncharacterized protein LOC105650827 [Jatropha curcas] |