| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
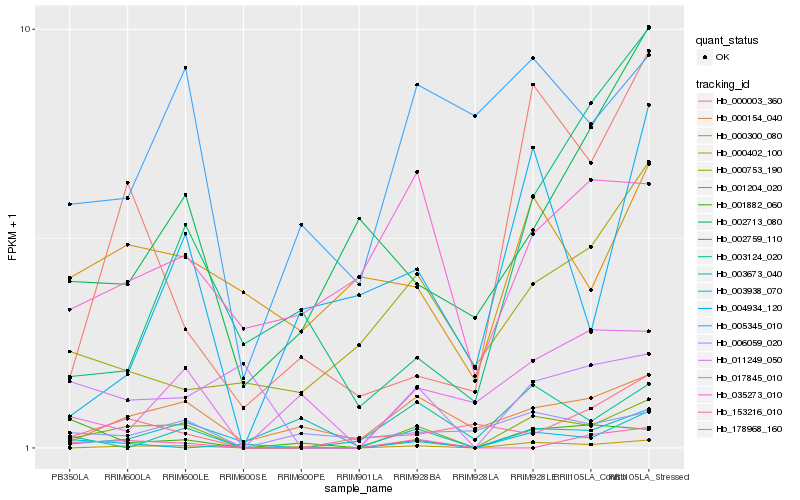
Hb_001204_020 |
0.0 |
- |
- |
Uncharacterized protein TCM_021965 [Theobroma cacao] |
| 2 |
Hb_000402_100 |
0.2396382619 |
- |
- |
PREDICTED: L-tryptophan--pyruvate aminotransferase 1 [Jatropha curcas] |
| 3 |
Hb_000154_040 |
0.2994605318 |
- |
- |
hypothetical protein JCGZ_25602 [Jatropha curcas] |
| 4 |
Hb_000003_360 |
0.3026835127 |
- |
- |
Putative inactive purple acid phosphatase 27 [Glycine soja] |
| 5 |
Hb_003124_020 |
0.3222628083 |
transcription factor |
TF Family: NAC |
PREDICTED: uncharacterized protein LOC105111046 [Populus euphratica] |
| 6 |
Hb_011249_050 |
0.3314293813 |
- |
- |
- |
| 7 |
Hb_153216_010 |
0.3351166484 |
- |
- |
PREDICTED: uncharacterized protein LOC105797726 [Gossypium raimondii] |
| 8 |
Hb_017845_010 |
0.339197971 |
- |
- |
- |
| 9 |
Hb_003938_070 |
0.3424357715 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 10 |
Hb_006059_020 |
0.3449337233 |
- |
- |
PREDICTED: uncharacterized protein LOC105648672 [Jatropha curcas] |
| 11 |
Hb_178968_160 |
0.3449960457 |
- |
- |
- |
| 12 |
Hb_005345_010 |
0.3470588636 |
- |
- |
PREDICTED: telomere repeat-binding factor 4-like [Jatropha curcas] |
| 13 |
Hb_001882_060 |
0.3490459188 |
- |
- |
- |
| 14 |
Hb_002713_080 |
0.3507881136 |
- |
- |
glycine-rich RNA-binding protein, putative [Ricinus communis] |
| 15 |
Hb_002759_110 |
0.3555652466 |
- |
- |
RNA-binding family protein, putative isoform 1 [Theobroma cacao] |
| 16 |
Hb_000753_190 |
0.3582386481 |
- |
- |
PREDICTED: D-aminoacyl-tRNA deacylase [Populus euphratica] |
| 17 |
Hb_035273_010 |
0.359149735 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647563 [Jatropha curcas] |
| 18 |
Hb_004934_120 |
0.3599133255 |
- |
- |
- |
| 19 |
Hb_000300_080 |
0.3611974571 |
- |
- |
PREDICTED: probable S-sulfocysteine synthase, chloroplastic isoform X2 [Jatropha curcas] |
| 20 |
Hb_003673_040 |
0.3623141035 |
- |
- |
dynamin family protein [Populus trichocarpa] |