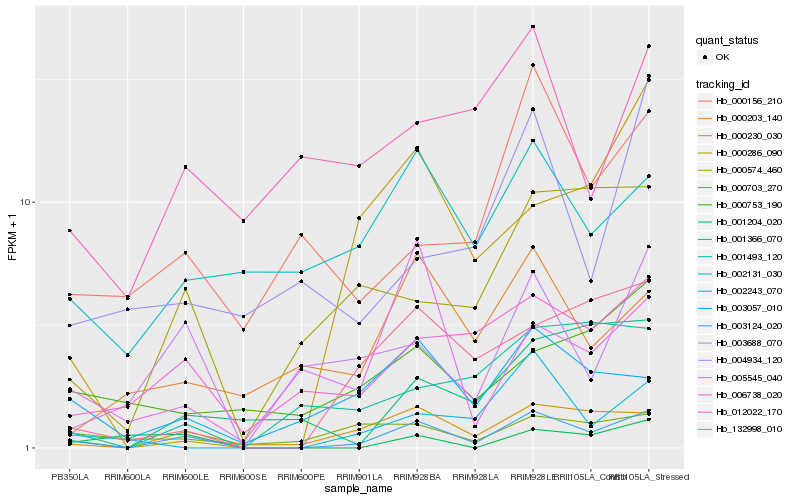
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003124_020 |
0.0 |
transcription factor |
TF Family: NAC |
PREDICTED: uncharacterized protein LOC105111046 [Populus euphratica] |
| 2 |
Hb_000230_030 |
0.2233971766 |
- |
- |
hypothetical protein B456_013G2021001, partial [Gossypium raimondii] |
| 3 |
Hb_003057_010 |
0.2699669444 |
- |
- |
hypothetical protein JCGZ_06666 [Jatropha curcas] |
| 4 |
Hb_132998_010 |
0.272399653 |
- |
- |
hypothetical protein JCGZ_00197 [Jatropha curcas] |
| 5 |
Hb_006738_020 |
0.2771012601 |
- |
- |
hypothetical protein VITISV_010510 [Vitis vinifera] |
| 6 |
Hb_000574_460 |
0.2842423442 |
- |
- |
hypothetical protein JCGZ_06119 [Jatropha curcas] |
| 7 |
Hb_002243_070 |
0.2911292657 |
- |
- |
sodium-bile acid cotransporter, putative [Ricinus communis] |
| 8 |
Hb_004934_120 |
0.2929070231 |
- |
- |
- |
| 9 |
Hb_000286_090 |
0.304634423 |
transcription factor |
TF Family: B3 |
hypothetical protein JCGZ_00197 [Jatropha curcas] |
| 10 |
Hb_001366_070 |
0.3050577864 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 4-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_000156_210 |
0.3064944163 |
- |
- |
PREDICTED: metal transporter Nramp1 [Jatropha curcas] |
| 12 |
Hb_001493_120 |
0.3088469546 |
- |
- |
- |
| 13 |
Hb_002131_030 |
0.3088920294 |
- |
- |
PREDICTED: pyruvate kinase isozyme G, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000703_270 |
0.3105786245 |
- |
- |
hypothetical protein B456_012G039600 [Gossypium raimondii] |
| 15 |
Hb_000753_190 |
0.3131530577 |
- |
- |
PREDICTED: D-aminoacyl-tRNA deacylase [Populus euphratica] |
| 16 |
Hb_000203_140 |
0.3143777724 |
- |
- |
PREDICTED: peroxisome biogenesis protein 7 [Jatropha curcas] |
| 17 |
Hb_005545_040 |
0.3153264494 |
- |
- |
PREDICTED: protein LURP-one-related 14 isoform X2 [Nelumbo nucifera] |
| 18 |
Hb_001204_020 |
0.3222628083 |
- |
- |
Uncharacterized protein TCM_021965 [Theobroma cacao] |
| 19 |
Hb_003688_070 |
0.3227298246 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein E [Jatropha curcas] |
| 20 |
Hb_012022_170 |
0.3283056932 |
transcription factor |
TF Family: HMG |
DNA-binding protein MNB1B, putative [Ricinus communis] |