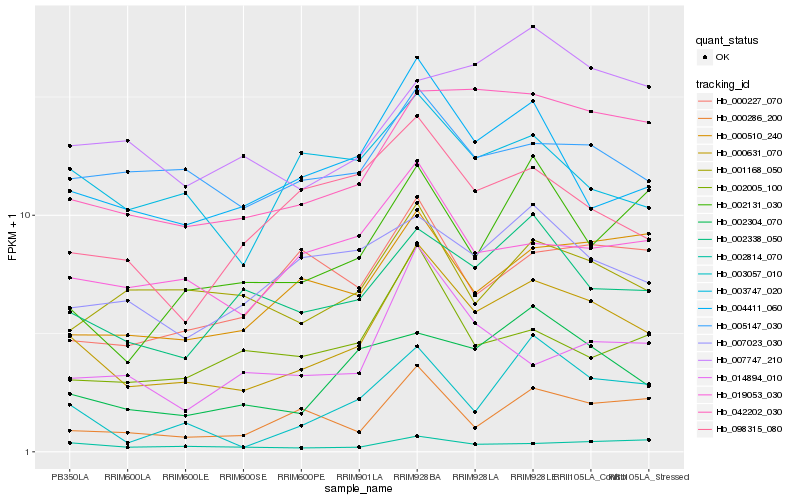
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000631_070 |
0.0 |
transcription factor |
TF Family: C2H2 |
PREDICTED: histone deacetylase 5 [Jatropha curcas] |
| 2 |
Hb_042202_030 |
0.1277906379 |
- |
- |
PREDICTED: uncharacterized protein LOC105647255 [Jatropha curcas] |
| 3 |
Hb_004411_060 |
0.1397467179 |
- |
- |
acetolactate synthase, putative [Ricinus communis] |
| 4 |
Hb_002338_050 |
0.1435826958 |
- |
- |
PREDICTED: uncharacterized protein LOC105118489 isoform X1 [Populus euphratica] |
| 5 |
Hb_002304_070 |
0.1458662956 |
- |
- |
- |
| 6 |
Hb_019053_030 |
0.1505627926 |
- |
- |
PREDICTED: sulfite oxidase [Jatropha curcas] |
| 7 |
Hb_002814_070 |
0.1543431256 |
- |
- |
PREDICTED: uncharacterized protein LOC104877459 [Vitis vinifera] |
| 8 |
Hb_000510_240 |
0.1544784762 |
desease resistance |
Gene Name: PRK |
ATP binding protein, putative [Ricinus communis] |
| 9 |
Hb_000286_200 |
0.1606377909 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X2 [Jatropha curcas] |
| 10 |
Hb_007023_030 |
0.1619153785 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g20710, mitochondrial-like [Jatropha curcas] |
| 11 |
Hb_005147_030 |
0.1628292834 |
- |
- |
hypothetical protein POPTR_0004s23390g [Populus trichocarpa] |
| 12 |
Hb_098315_080 |
0.163011825 |
- |
- |
methylmalonate-semialdehyde dehydrogenase, putative [Ricinus communis] |
| 13 |
Hb_003747_020 |
0.1654119944 |
- |
- |
PREDICTED: acetyl-coenzyme A synthetase, chloroplastic/glyoxysomal isoform X1 [Jatropha curcas] |
| 14 |
Hb_002005_100 |
0.1661746155 |
- |
- |
- |
| 15 |
Hb_001168_050 |
0.1668016689 |
- |
- |
hypothetical protein POPTR_0007s03190g [Populus trichocarpa] |
| 16 |
Hb_002131_030 |
0.1677006801 |
- |
- |
PREDICTED: pyruvate kinase isozyme G, chloroplastic [Jatropha curcas] |
| 17 |
Hb_003057_010 |
0.1680270323 |
- |
- |
hypothetical protein JCGZ_06666 [Jatropha curcas] |
| 18 |
Hb_014894_010 |
0.1699235438 |
- |
- |
hypothetical protein CICLE_v10001869mg [Citrus clementina] |
| 19 |
Hb_000227_070 |
0.1702287895 |
- |
- |
PREDICTED: ATP-dependent (S)-NAD(P)H-hydrate dehydratase isoform X2 [Jatropha curcas] |
| 20 |
Hb_007747_210 |
0.170239709 |
- |
- |
unnamed protein product [Vitis vinifera] |